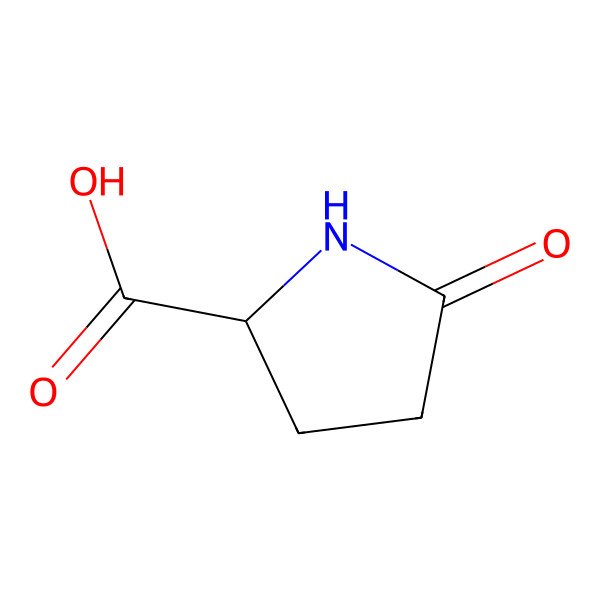
| 98-79-3 |
| Pidolic acid |
| Pyroglutamic acid |
| 5-OXOPROLINE |
| 5-oxo-L-proline |
| H-Pyr-OH |
| (S)-5-Oxopyrrolidine-2-carboxylic Acid |
| (2S)-5-oxopyrrolidine-2-carboxylic acid |
| Pyroglutamate |
| pyroglu |
| GLUTIMIC ACID |
| L-Proline, 5-oxo- |
| (S)-(-)-2-Pyrrolidone-5-carboxylic acid |
| 5-L-Oxoproline |
| Pyrrolidonecarboxylic acid |
| L-5-Oxoproline |
| Pidolate |
| Pidolidone |
| Proline, 5-oxo- |
| (-)-2-Pyrrolidone-5-carboxylic acid |
| AJIDEW A-100 |
| (S)-PYROGLUTAMIC ACID |
| 5-Carboxy-2-pyrrolidinone |
| OXOPROLINE |
| L-Pyroglutamate |
| 2-Pyrrolidinone-5-carboxylic acid |
| L-5-Carboxy-2-pyrrolidinone |
| L-GLUTIMINIC ACID |
| L-5-Oxo-2-pyrrolidinecarboxylic acid |
| (S)-5-Oxo-2-pyrrolidinecarboxylic acid |
| 5-Pyrrolidinone-2-carboxylic acid |
| Pyr-OH |
| 2-L-Pyrrolidone-5-carboxylic acid |
| GLUTIMINIC ACID |
| Pidolic acid [INN] |
| Pyroglutamic acid, l- |
| L-GLUTIMIC ACID |
| oxo-proline |
| Acide pidolique |
| Acido pidolico |
| 5-Pyrrolidone-2-carboxylic acid |
| Acidum pidolicum |
| L-2-PYRROLIDONE-5-CARBOXYLIC ACID |
| L-5-Pyrrolidone-2-carboxylic acid |
| L-pca |
| 2-Pyrrolidone-5-carboxylate |
| l-pyrrolidone carboxylic acid |
| L-Glutamic acid gamma-lactam |
| NSC 143034 |
| Acide pidolique [INN-French] |
| Acido pidolico [INN-Spanish] |
| Acidum pidolicum [INN-Latin] |
| (5S)-2-Oxopyrrolidine-5-carboxylic acid |
| 5-OXO-2-PYRROLIDINECARBOXYLIC ACID |
| PYRROLIDINONECARBOXYLIC ACID |
| L-PYRROLIDONECARBOXYLIC ACID |
| 2-Oxopyrrolidine-5-carboxylic acid |
| NSC 9966 |
| EINECS 202-700-3 |
| L-PYRROLIDINONECARBOXYLIC ACID |
| PYRROLIDONE-5-CARBOXYLIC ACID |
| Proline, 5-oxo-, L- |
| Pidolic acid [INN:BAN] |
| 2-Benzothiazolesulfenic acid morpholide |
| NSC-760414 |
| UNII-SZB83O1W42 |
| SZB83O1W42 |
| DTXSID6046260 |
| CHEBI:18183 |
| NSC9966 |
| (S)-2-Pyrrolidone-5-carboxylic acid |
| NSC-143034 |
| NCGC00160613-01 |
| DTXCID4026260 |
| CAS-98-79-3 |
| pGlu-OH |
| SMR000857158 |
| Glutiminate |
| Glutimate |
| pidolic-acid |
| pidolinsyre- |
| pGlu |
| L-Glutiminate |
| L-Glutimate |
| (S)-Pyroglutamate |
| (-)-Pyroglutamate |
| glutamic acid lactam |
| MFCD00005272 |
| L-pyro-glutamic acid |
| Pyrrolidinonecarboxylate |
| (L)-pyroglutamic acid |
| Ajidew A 100 |
| L-Pyrrolidonecarboxylate |
| Oxopyrrolidinecarboxylate |
| (-)-Pyroglutamic acid |
| L-Glutamic Acid Lactam |
| L-Pyrrolidinonecarboxylate |
| Pyrrolidone-5-carboxylate |
| L-Glutamic acid g-lactam |
| PCA [INCI] |
| Rec-PGA 273K |
| pyrrolidone car-boxylic acid |
| L-Pyroglutamic acid, 97% |
| Oxopyrrolidinecarboxylic acid |
| SCHEMBL15790 |
| (S)-(-)-pyroglutamic acid |
| L-Pyroglutamic acid, BioXtra |
| MLS001332421 |
| MLS001332422 |
| 2-Pyrrolidinone-5-carboxylate |
| 5-Pyrrolidinone-2-carboxylate |
| alpha-aminoglutaric acid lactam |
| 2-L-Pyrrolidone-5-carboxylate |
| L-2-Pyrrolidone-5-carboxylate |
| PIDOLIC ACID [WHO-DD] |
| CHEMBL397976 |
| L-Glutamic acid .gamma.-lactam |
| PYROGLUTAMIC ACID, (L) |
| L-PYROGLUTAMIC ACID [MI] |
| (S)-2-Pyrrolidone-5-carboxylate |
| L-5-Oxo-2-pyrrolidinecarboxylate |
| (-)-2-Pyrrolidone-5-carboxylate |
| HMS2231L11 |
| HMS3264E20 |
| Pharmakon1600-01506185 |
| 2-Oxopyrrolidine-5(S)-carboxylate |
| PYROGLUTAMIC ACID [USP-RS] |
| CS-M0659 |
| GLP |
| NSC-9966 |
| NSC11742 |
| STR02331 |
| (S)-5-Oxo-2-pyrrolidinecarboxylate |
| Tox21_111936 |
| CCG-36432 |
| GEO-04256 |
| NSC-11742 |
| NSC760414 |
| s5823 |
| (5S)-2-Oxopyrrolidine-5-carboxylate |
| AKOS015855330 |
| Tox21_111936_1 |
| (2S)-5-pyrrolidone-2-carboxylic acid |
| (S)-(-)-g-Butyrolactam-g-carboxylate |
| (S)-5-oxopyrolidine-2-carboxylic acid |
| 2-Oxopyrrolidine-5(S)-carboxylic acid |
| AM83732 |
| DB03088 |
| (S)-(-)-2-Pyrrolidone-5-carboxylate |
| L-Pyroglutamic acid, >=99.0% (T) |
| (5S)-2-xopyrrolidine-5-carboxylic acid |
| NCGC00160613-02 |
| (S)-5-oxo-pyrrolidine-2-carboxylic acid |
| AC-15173 |
| BP-12844 |
| HY-76082 |
| (S)-(-)-g-Butyrolactam-g-carboxylic acid |
| LS-118965 |
| (s)-(-)-2-pyrrolidinone-5-carboxylic acid |
| P0573 |
| (S)-(-)-gamma-Butyrolactam-gamma-carboxylate |
| EN300-72896 |
| C01879 |
| M03204 |
| P-8490 |
| P17107 |
| AB00514366_07 |
| (S)-(-)-gamma-Butyrolactam-gamma-carboxylic acid |
| A845910 |
| Q60998677 |
| F8889-8712 |
| Z1154378309 |
| Pidolic acid, European Pharmacopoeia (EP) Reference Standard |
| E478F48D-E369-43AE-8132-08D819242518 |
| Pyroglutamic Acid, United States Pharmacopeia (USP) Reference Standard |
| 35255-51-7 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|