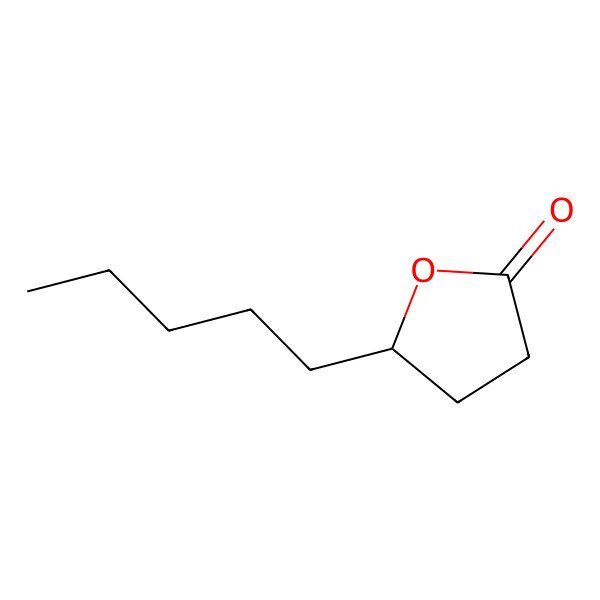
| Gamma-nonalactone |
| gamma-Nonanolactone |
| 5-pentyldihydrofuran-2(3H)-one |
| 5-pentyloxolan-2-one |
| 2(3H)-Furanone, dihydro-5-pentyl- |
| Prunolide |
| gamma-Nonanolide |
| Nonan-1,4-olide |
| Cocos aldehyde |
| Coconut aldehyde |
| 4-Nonanolide |
| 1,4-Nonalolide |
| 4-Pentyl-butanolide |
| Aldehyde C-18 |
| 4-Nonalactone |
| DIHYDRO-5-PENTYL-2(3H)-FURANONE |
| gamma-Nonylactone |
| FEMA No. 2781 |
| gamma-Nonyllactone |
| Nonan-4-olide |
| gamma-Pelargolactone |
| 1,4-Nonyl lactone |
| gamma-Nonanoic lactone |
| delta-n-Amylbutyrolactone |
| FEMA Number 2781 |
| gamma-N-Amylbutyrolactone |
| gamma-Amyl-gamma-butyrolactone |
| .gamma.-Nonalactone |
| gamma-Pentyl-gamma-butyrolactone |
| HSDB 5360 |
| rac-gamma-Nonanolactone |
| EINECS 203-219-1 |
| 4-Amyl-4-hydroxybutyric acid lactone |
| 4-Hydroxynonanoic acid, gamma-lactone |
| UNII-I1XGH66S8P |
| .gamma.-n-Amylbutyrolactone |
| I1XGH66S8P |
| Nonanoic acid, 4-hydroxy-, gamma-lactone |
| AI3-02457 |
| CHEMBL191935 |
| DTXSID0034229 |
| Dihydro-5-pentylfuran-2(3H)-one |
| 4-Hydroxynonanoic acid gamma-lactone |
| NSC 24253 |
| NSC 46099 |
| NSC-24253 |
| NSC-46099 |
| apricolin |
| 4-Hydroxynonanoic acid, .gamma.-lactone |
| EC 203-219-1 |
| 82373-92-0 |
| 4-Pentylbutanolide |
| .gamma.-Nonalacton |
| .gamma.-Nonanolide |
| ?-Nonanoic lactone |
| Nonan- 4- olide |
| MFCD00005403 |
| gamma-Pelargonolactone |
| .gamma.-Nonanolactone |
| NONYL LACTONE |
| nonan - 4 - olide |
| gamma -Nonanoic lactone |
| (+-)-gamma-Nonalactone |
| .gamma.-Amylbutyrolactone |
| .gamma.-Nonanoic lactone |
| 5-Pentyl-.gamma.-lactone |
| gamma-Nonanoic lactone, 97% |
| SCHEMBL112484 |
| 4-Hydroxynonanoic acid lactone |
| 5-Pentyl-dihydro-furan-2-one |
| GAMMA-NONALACTONE [FCC] |
| DTXCID8014229 |
| GAMMA-NONALACTONE [INCI] |
| 5-pentyl-tetrahydro-furan-2-one |
| OALYTRUKMRCXNH-UHFFFAOYSA- |
| (.+/-.)-.gamma.-Nonalactone |
| 5-Pentyldihydro-2(3H)-furanone |
| Dihydro-5-pentyl-2(3H)-furanon |
| .gamma.-Amyl-.gamma.-butyrolactone |
| .GAMMA.-NONALACTONE [FHFI] |
| NSC24253 |
| NSC46099 |
| (3H)-Dihydro-5-pentyl-2-furanone |
| 5-Pentyldihydro-2(3H)-furanone # |
| Tox21_301039 |
| BBL027520 |
| BDBM50167995 |
| LMFA07040021 |
| STL373781 |
| AKOS015904806 |
| CS-W017997 |
| DS-3247 |
| LS-2984 |
| DELTA-N-AMYLBUTYROLACTONE [HSDB] |
| NCGC00164129-01 |
| NCGC00164129-02 |
| NCGC00254941-01 |
| CAS-104-61-0 |
| gamma-Nonanoic lactone, >=98%, FCC, FG |
| gamma-Nonanoic lactone, natural, 98%, FG |
| FT-0686718 |
| N0285 |
| Nonanoic acid, 4-hydroxy-, .gamma.-lactone |
| D70273 |
| EN300-342288 |
| gamma-Nonalactone, analytical reference material |
| J-001202 |
| Q11255995 |
| Z1255430984 |
| InChI=1/C9H16O2/c1-2-3-4-5-8-6-7-9(10)11-8/h8H,2-7H2,1H3 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|