| colcemid |
| 477-30-5 |
| Colchamine |
| (-)-Demecolcine |
| Demecolcin |
| Reichstein's F |
| Colcemide |
| Desmecolcine |
| Substance F |
| N-Deacetyl-N-methylcolchicine |
| Santavy's substance F |
| (-)-Colchamine |
| Kolchamin |
| Kolkamin |
| Omain |
| Omaine |
| Methylcolchicine |
| N-Methyl-N-deacetylcolchicine |
| Deacetyl-N-methylcolchicine |
| Deacetylmethylcolchicine |
| N-Desacetyl-N-methylcolchicine |
| N-Methyl-N-desacetylcolchicine |
| Desacetylmethylcolchicine |
| Demecolcina |
| Demecolcinum |
| Kolchicin |
| Kolchicin [Czech] |
| N-Desacetylmethylcolchicine |
| Colchicine, N-deacetyl-N-methyl- |
| Colchamin |
| Alkaloid H 3, from colchicum antumnale |
| C-12669 |
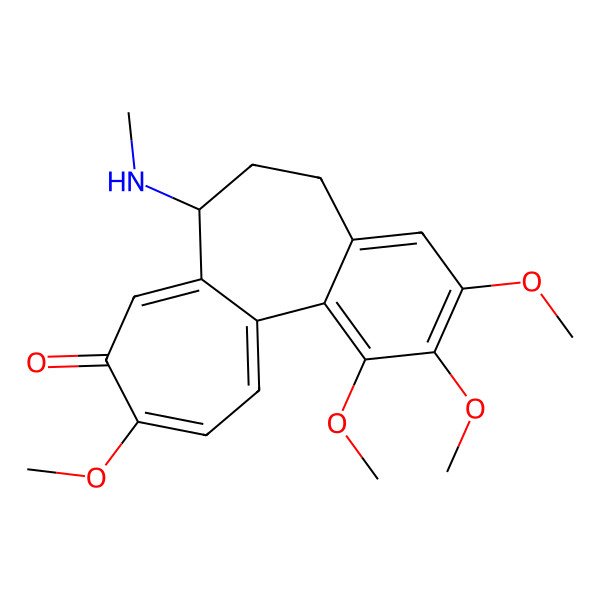
| (S)-1,2,3,10-Tetramethoxy-7-(methylamino)-6,7-dihydrobenzo[a]heptalen-9(5H)-one |
| Demecolcinum [INN-Latin] |
| Ciba 12669A |
| Demecolcina [INN-Spanish] |
| NSC-3096 |
| Colchicine, 7-deacetamido-7-(methylamino)- |
| X 153 |
| Demecolcine [INN:BAN:DCF] |
| CCRIS 2764 |
| C21H25NO5 |
| (7S)-1,2,3,10-tetramethoxy-7-(methylamino)-6,7-dihydrobenzo[a]heptalen-9(5H)-one |
| NSC 3096 |
| EINECS 207-514-6 |
| UNII-Z01IVE25KI |
| BRN 2822892 |
| Z01IVE25KI |
| Ciba 12669 A |
| (7S)-1,2,3,10-tetramethoxy-7-(methylamino)-6,7-dihydro-5H-benzo[a]heptalen-9-one |
| CHEBI:4393 |
| CHEMBL312862 |
| Colchicine, deacetyl-N-methyl- |
| DTXSID7020342 |
| Benzo(a)heptalen-9(5H)-one, 6,7-dihydro-1,2,3,10-tetramethoxy-7-(methylamino)-, (S)- |
| 6,7-Dihydro-1,2,3,10-tetramethoxy-7-(methylamino)-benzo(alpha)heptalen-9(5H)-one |
| Demicolcine |
| (7S)-7-(methylamino)-1,2,3,10-tetrakis(methyloxy)-6,7-dihydrobenzo[a]heptalen-9(5H)-one |
| SMR000058249 |
| 4-14-00-00940 (Beilstein Handbook Reference) |
| Demecolcine [BAN:DCF:INN] |
| WLN: L B677 MV&T&J CO1 DO1 EO1 JM1 NO1 |
| NSC3096 |
| Desmecolchine |
| N-de-nu-acetyl-N-methylcolchicine |
| Benzo[a]heptalen-9(5H)-one, 6,7-dihydro-1,2,3,10-tetramethoxy-7-(methylamino)-, (S)- |
| Benzo[a]heptalen-9(5H)-one,7-dihydro-1,2,3,10-tetramethoxy-7-(methylamino)-, (S)- |
| Colcemid;Colchamin;Desmecolcine;Kolchamin |
| NCGC00166035-01 |
| Alkaloid H 3 |
| MFCD00075459 |
| DEMECOLCINE [MI] |
| DEMECOLCINE [INN] |
| SCHEMBL8161 |
| DEMECOLCINE [WHO-DD] |
| MLS001332501 |
| MLS001332502 |
| MLS002695919 |
| DTXCID70342 |
| Colchine, N-deacetyl-N-methyl |
| cid_220401 |
| BCBcMAP01_000022 |
| Demecolcine, >=98% (HPLC) |
| (7S)-6,7-Dihydro-1,2,3,10-tetramethoxy-7-(methylamino)benzo[a]heptalen-9(5H)-one |
| Benzo[a]heptalen-9(5H)-one, 6,7-dihydro-1,2,3,10-tetramethoxy-7-(methylamino)-, (7S)- |
| NNJPGOLRFBJNIW-HNNXBMFYSA-N |
| HMS2230L17 |
| HY-N0282 |
| Tox21_112296 |
| BDBM50014872 |
| NSC403147 |
| AKOS002141248 |
| DB13318 |
| KS-5002 |
| LS-7285 |
| NSC-403147 |
| SMP1_000076 |
| Colcemid, N-methyl-N-deacetyl-colchicine |
| NCGC00166035-02 |
| CAS-477-30-5 |
| 4-Amino-furazan-3-carboxylicacidmethylamide |
| CS-0008779 |
| A827317 |
| Q903666 |
| SR-01000841244 |
| Demecolcine, >=98.0% (sum of enantiomers, HPLC) |
| SR-01000841244-3 |
| BRD-K38624570-001-10-7 |
| 6,2,3,10-tetramethoxy-7-(methylamino)-benzo[.alpha.]heptalen-9(5H)-one |
| Demecolcine, Hybri-Max(TM), powder, gamma-irradiated, hybridoma tested |
| (S)-1,2,3,10-tetramethoxy-7-methylamino-6,7-dihydro-5H-benzo[a]heptalen-9-one |
| (S)-6,7-dihydro-1,2,3,10-tetramethoxy-7-(methylamino)benzo[a]heptalen-9(5H)-one |
| 1,2,3,10-Tetramethoxy-7-(methylamino)-6,7-dihydrobenzo[a]heptalen-9(5H)-one # |
| 1,2,3,10-Tetramethoxy-7-methylamino-6,7-dihydro-5H-benzo[a]heptalen-9-one (Demecolcine) |
| 6,7-Dihydro-1,2,3,10-tetramethoxy-7-(methylamino)-benzo(a)heptalen-9(5H)-one |
| LON |
|
There are more than 10 synonyms. If you wish to see them all click here.
|