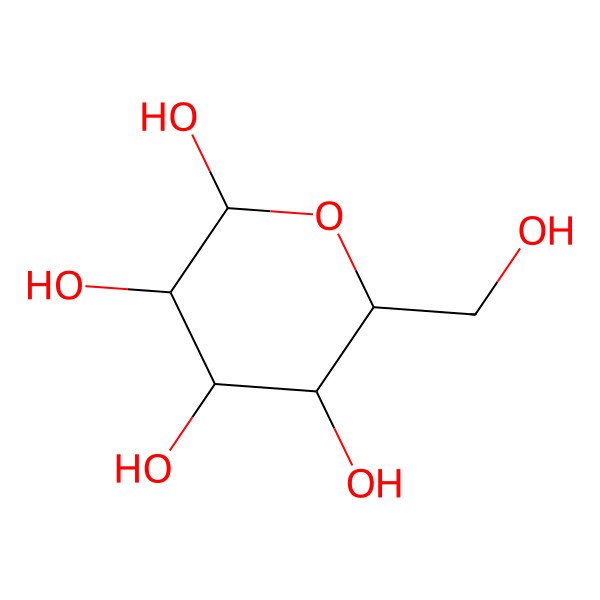
| beta-D-glucopyranose |
| 492-61-5 |
| glucoside |
| beta-Dextrose |
| b-d-glucose |
| .beta.-D-Glucopyranose |
| beta-glucose |
| Curdlan |
| 28905-12-6 |
| Oxidase, glucose |
| (2R,3R,4S,5S,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrol |
| .beta.-D-Glucose |
| Glucose, (beta-D)-Isomer |
| (2R,3R,4S,5S,6R)-6-(Hydroxymethyl)tetrahydro-2H-pyran-2,3,4,5-tetraol |
| .beta.-d-Glucose, anhydrous |
| UNII-J4R00M814D |
| Grape sugar |
| beta-D-Glucopyranose, anhydrous |
| CHEMBL1614854 |
| CHEBI:15903 |
| 9001-37-0 |
| J4R00M814D |
| EINECS 207-756-2 |
| (+)-Glucose |
| 4-Morpholineacetic acid, a-Methylene-, Methyl ester |
| 128009-02-9 |
| 133947-06-5 |
| 54724-00-4 |
| ZYMOSAN |
| Glucodin |
| Meritose |
| 136760-05-9 |
| anhydrous glucose |
| Clintose L |
| 9010-72-4 |
| CPC hydrate |
| Roferose ST |
| Clearsweet 95 |
| Glucosides |
| Staleydex 95M |
| .beta.-D-ribo-Hexopyranose, 1,6-anhydro-3-deoxy-2-O-phenyl-4-O-(phenylmethyl)- |
| BGC |
| mikrotsid |
| Corylophyline |
| Callose |
| Microcid |
| Notatin |
| beta -d-glucose |
| beta-D-Glc |
| Deoxin-1 |
| 1,3-beta-D-Glucan |
| beta-(1,3)-glucan |
| beta-D-Glucose oxidase |
| I(2)-D-Glucopyranose |
| Glucose aerodehydrogenase |
| Beta-D-glucose anhydrous |
| Glucose oxidase [USAN] |
| Beta-d-glucose, anhydrous |
| 6-(hydroxymethyl)tetrahydro-2H-pyran-2,3,4,5-tetrol |
| GLUCOSE, BETA-D- |
| D07ONX |
| (1,2-beta-D-glucosyl)n |
| (1->2)-beta-D-glucan |
| (1->3)-beta-D-glucan |
| (1->4)-beta-D-glucan |
| (1->6)-beta-D-glucan |
| GLUCOSE, .BETA.-D |
| SCHEMBL25601 |
| (1->2)-beta-D-glucopyranan |
| (1->3)-beta-D-glucopyranan |
| (1->4)-beta-D-glucopyranan |
| (1->6)-beta-D-glucopyranan |
| CCRIS 3425 |
| UNII-0T8392U5N1 |
| CHEBI:18246 |
| CHEBI:27380 |
| CHEBI:27517 |
| CHEBI:37671 |
| DTXSID70883403 |
| Pharmakon1600-01300015 |
| .BETA.-D-GLUCOSE ANHYDROUS |
| beta-D-glucose; D-glucose; glucose |
| EINECS 232-601-0 |
| BDBM50240803 |
| beta-D-Glucose:quinone oxidoreductase |
| MFCD00063989 |
| NSC759603 |
| beta-D-Glucopyranose aerodehydrogenase |
| AKOS016010209 |
| DB02379 |
| NSC-759603 |
| YC46078 |
| beta-D-glucose: oxygen 1-oxidoreductase |
| ?-D-Glucose (contains alpha-D-Glucose) |
| NCGC00263446-02 |
| Beta-D-glucose(contains alpha-D-glucose) |
| BS-22220 |
| HY-121965 |
| CS-0083772 |
| G0047 |
| C00221 |
| D90709 |
| E.C. 1.1.3.4 |
| AE-562/43459286 |
| W-202206 |
| Glucose oxidase (EC 1.1.3.4) from aspergillus niger |
| Q23905968 |
| 50986-29-3 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|