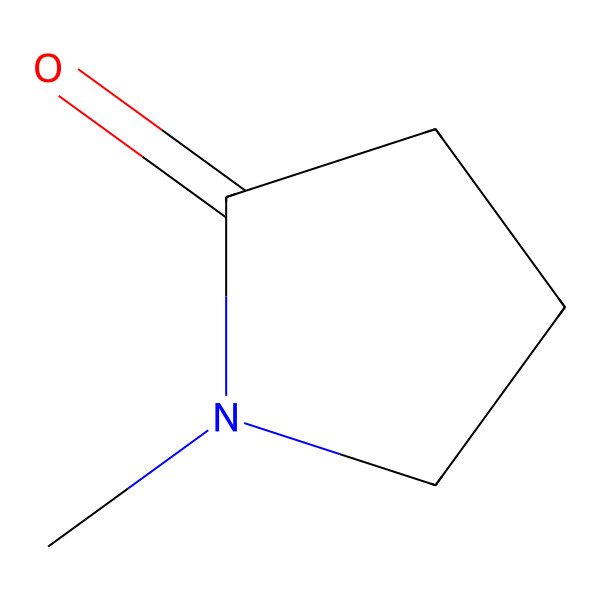
| 872-50-4 |
| N-Methylpyrrolidone |
| N-Methyl-2-pyrrolidone |
| 1-methylpyrrolidin-2-one |
| Methylpyrrolidone |
| 1-Methyl-2-pyrrolidone |
| N-Methyl-2-pyrrolidinone |
| M-Pyrol |
| 1-Methylpyrrolidinone |
| Methyl pyrrolidone |
| 1-Methylpyrrolidone |
| N-Methylpyrrolidinone |
| n-methyl-pyrrolidone |
| 2-Pyrrolidinone, 1-methyl- |
| 1-Methyl-5-pyrrolidinone |
| 1-Methylazacyclopentan-2-one |
| N-methylpyrrolidin-2-one |
| NMP |
| N-Methyl-gamma-butyrolactam |
| N-methyl pyrrolidone |
| 1-methylpyrrolidine-2-one |
| N-methyl pyrrolidinone |
| Methyl-2-pyrrolidinone |
| 1-methyl-2-pyrrolidon |
| Methylpyrrolidinone |
| 2-Pyrrolidinone, methyl- |
| 2687-44-7 |
| Methylpyrrolidone [NF] |
| N-Methylpyrrolid-2-one |
| N-Methyl-alpha-pyrrolidone |
| NSC 4594 |
| Agsolex 1 |
| N-Methyl-alpha-pyrrolidinone |
| CCRIS 1633 |
| 51013-18-4 |
| DTXSID6020856 |
| Methylpyrrolidone, N- |
| HSDB 5022 |
| Pyrrolidinone, methyl- |
| NSC-4594 |
| EINECS 212-828-1 |
| UNII-JR9CE63FPM |
| MFCD00003193 |
| JR9CE63FPM |
| 1-methyl-pyrrolidin-2-one |
| Norleucine, 5-oxo-, DL- |
| N-Methyl-.alpha.-pyrrolidone |
| 1-Methyl-2-pyrrolidinone-d9 |
| CHEMBL12543 |
| N-Methyl-.gamma.-butyrolactam |
| AI3-23116 |
| CHEBI:7307 |
| N-Methyl-.alpha.-pyrrolidinone |
| 1-Methyl-2-pyrrolidinone, anhydrous |
| 1-Methyl-2-pyrrolidinone, HPLC Grade |
| EC 212-828-1 |
| N 0131 |
| 30207-69-3 |
| DTXCID60856 |
| pharmasolve |
| N-Methylpyrrolidon |
| CAS-872-50-4 |
| N-methyl-pyrrolidinone |
| N-Methyl-2-pyrrolidon |
| 1-methyl-2-pyrolidone |
| N-methyl-pyrrolidin-2-one |
| 1-Methyl-2-pyrrolidinone, puriss. p.a., >=99.0% (GC) |
| Micropure ultra |
| N-methylpyrolidone |
| N-methypyrrolidone |
| Max-1 peptide |
| Pyrol M |
| N-methylpirrolidone |
| 1methylpyrrolidinone |
| n-methyl pyrrolidon |
| N-methy pyrrolidone |
| N-methyl-pyrolidone |
| N-methyl-pyrrolidon |
| N-methylpyrolidinone |
| 1-methylpyrolidinone |
| Microposit 2001 |
| n-methylpyrollidinone |
| N-Methylpyrrolidione |
| N-methlypyrrolidinone |
| N-methyl pirrolidone |
| N-methyl pyrollidone |
| N-methyl-pyrollidone |
| N-methylpyrrolidone- |
| NMP,SP Grade |
| 1-methyl pyrrolidone |
| 1-methyl-pyrrolidone |
| methyl-2-pyrrolidone |
| N-methy pyrrolidinone |
| N-methyl pyrolidinone |
| N-methyl-pyrolidinone |
| N-methyl- pyrrolidone |
| N-methylpyrro-lidinone |
| N-methylpyrroli-dinone |
| N-methylpyrrolidin-one |
| 1-methyl-2pyrrolidone |
| 1-methyl2-pyrrolidone |
| 1methyl-2-pyrrolidone |
| N-Metyl-2-pyrrolidon |
| 1-methyl pyrrolidinone |
| 1-methyl-pyrrolidinone |
| methylpyrrolidin-2-one |
| N-methy-2-pyrrolidone |
| N-methyl 2-pyrolidone |
| N-methyl-2-pyrolidone |
| 1-Metil-2-pirrolidona |
| 3p1d |
| N-methyl 2-pyrrolidone |
| N-methyl-2-pyrollidone |
| MPY (CHRIS Code) |
| 1-methyl-2-pirrolidone |
| 1-methyl-2-pyroldinone |
| 1-methylpyrrolid-2-one |
| 1methyl-2-pyrrolidinone |
| n-methylpyrrolidine-2one |
| N-methyl-2-pyrolidinone |
| N-methyl-2-pyrrolidinon |
| N-methylpyrolidin-2-one |
| 1-methy-2-pyrrolidinone |
| 1-methyl-2-pyrolidinone |
| N-methyl 2-pyrrolidinone |
| N-methyl-2-pyrollidinone |
| N-methyl-pyrrolid-2-one |
| N-methylpyrollidin-2-one |
| 1 -methyl-2-pyrrolidone |
| 1-methyl 2-pyrrolidinone |
| 1-methyl-2-pyrollidinone |
| 1-methyl-pyrrolin-2-one |
| N-Methylpyrrolidone-(2) |
| NMP, N-Methylpyrrolidone |
| 1-Methyl-pyrrolidin-2one |
| N-methylpyrrolidine-2-one |
| WLN: T5NVTJ A |
| N-methyl -2-pyrrolidinone |
| 1 -methyl-2-pyrrolidinone |
| 1-methyl -2-pyrrolidinone |
| 1-methyl-2- pyrrolidinone |
| 2-Pyrrolidone, 1-methyl- |
| 1-methyl-pyrrolidine-2-one |
| 2-pirrolidinona, 1-metil- |
| 1-N-methyl-2-pyrrolidinone |
| N-methyl-pyrrolidin -2-one |
| 1-Methylazacyclopentane-2-one |
| GTPL9520 |
| METHYL PYRROLIDONE [II] |
| 1-Methyl-2- pyrrolidin-2-one |
| N-Methyl-2-pyrrolidon (Dampf) |
| 1-METHYLPYRROLIDONE [MI] |
| METHYL PYRROLIDONE [INCI] |
| NSC4594 |
| METHYLPYRROLIDONE [USP-RS] |
| HY-Y1275 |
| N-METHYLPYRROLIDONE [MART.] |
| Tox21_202350 |
| Tox21_300097 |
| 1-Methyl-2-pyrrolidinone, 99.5% |
| BDBM50353587 |
| N-Methyl pyrrolidon (Peptide Grade) |
| N-METHYLPYRROLIDONE [USP-RS] |
| s6282 |
| STL183295 |
| N-Methyl-2-pyrrolidinone ACS reagent |
| AKOS000120930 |
| 1-Methyl-2-pyrrolidinone, BioSolv(R) |
| DB12521 |
| LS-1875 |
| SL 1332 |
| 1-Methyl-2-pyrrolidone, Reagent, ACS |
| 1-METHYL-2-PYRROLIDINONE [HSDB] |
| NCGC00247902-01 |
| NCGC00247902-02 |
| NCGC00253935-01 |
| NCGC00259899-01 |
| BP-31156 |
| N-METHYLPYRROLIDONE [EP MONOGRAPH] |
| LS-138890 |
| 1-Methyl-2-pyrrolidone (Low water content) |
| AM20110252 |
| CS-0017258 |
| FT-0608052 |
| FT-0672137 |
| FT-0698122 |
| FT-0700571 |
| M0418 |
| M3055 |
| 1-Methyl-2-pyrrolidinone, analytical standard |
| 1-Methyl-2-pyrrolidinone, anhydrous, 99.5% |
| 1-Methyl-2-pyrrolidinone, for HPLC, >=99% |
| 1-Methyl-2-pyrrolidinone, for synthesis, 99% |
| D78116 |
| M 0418 |
| Q33103 |
| Residual Solvent Class 2 - N-Methylpyrrolidone |
| 1-Methyl-2-pyrrolidinone, ReagentPlus(R), 99% |
| 1-Methyl-2-pyrrolidinone, Spectrophotometric Grade |
| 2-PYRROLIDONE,1-METHYL MFC5 H9 N1 O1 |
| A842053 |
| 1-Methyl-2-pyrrolidinone, ACS reagent, >=99.0% |
| 2,5-Dichloro-4,6-dimethyl pyridine-3-carbonitrile |
| J-504921 |
| J-803017 |
| Methyl 2-pyrrolidinone, 1-; (n-Methylpyrrolidone) |
| 1-Methyl-2-pyrrolidinone, biotech. grade, >=99.7% |
| 1-Methyl-2-pyrrolidinone, Electronic/Cleanroom Grade |
| 1-Methyl-2-pyrrolidinone, p.a., ACS reagent, 99% |
| 1-Methyl-2-pyrrolidone, anhydrous, water 40ppm max. |
| 1-Methyl-2-pyrrolidinone, SAJ first grade, >=98.0% |
| Z104478382 |
| 1-Methyl-2-pyrrolidinone, spectrophotometric grade, >=99% |
| 1-Methyl-2-pyrrolidinone, Vetec(TM) reagent grade, 98% |
| InChI=1/C5H9NO/c1-6-4-2-3-5(6)7/h2-4H2,1H |
| 2-PYRROLIDINONE, 1-METHYL-2,BETA-BUTOXYETHOXYETHYL CHLORIDE |
| 1-Methyl-2-pyrrolidinone, for metal speciation analysis, >=99.0% (GC) |
| Methylpyrrolidone, United States Pharmacopeia (USP) Reference Standard |
| N-Methylpyrrolidone, Pharmaceutical Secondary Standard; Certified Reference Material |
| 26876-92-6 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|