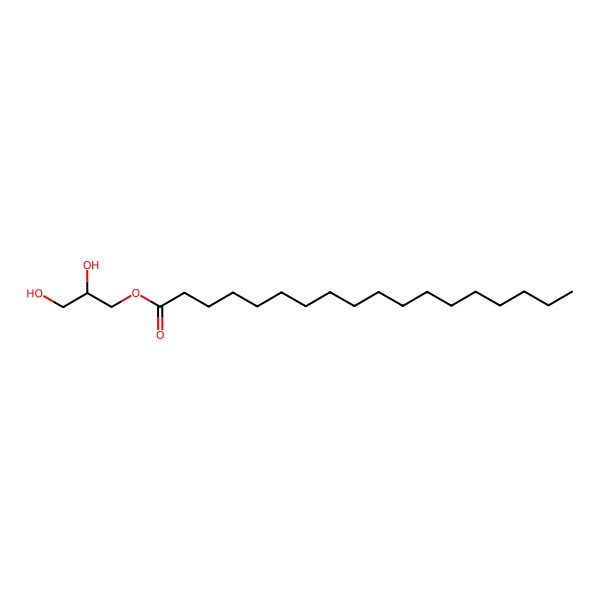
| 123-94-4 |
| Monostearin |
| 31566-31-1 |
| GLYCEROL MONOSTEARATE |
| Glyceryl stearate |
| Tegin |
| 1-Stearoyl-rac-glycerol |
| 1-MONOSTEARIN |
| Glycerin 1-monostearate |
| Stearin, 1-mono- |
| Stearic acid 1-monoglyceride |
| 2,3-dihydroxypropyl octadecanoate |
| Glycerol 1-monostearate |
| 1-Glyceryl stearate |
| Glycerin 1-stearate |
| Sandin EU |
| 1-Monostearoylglycerol |
| Octadecanoic acid, 2,3-dihydroxypropyl ester |
| Aldo MSD |
| Aldo MSLG |
| Glyceryl 1-monostearate |
| Stearoylglycerol |
| Glycerol 1-stearate |
| alpha-Monostearin |
| Tegin 55G |
| Emerest 2407 |
| Aldo 33 |
| Aldo 75 |
| Glycerin monostearate |
| Arlacel 165 |
| 3-Stearoyloxy-1,2-propanediol |
| Cerasynt SD |
| Stearin, mono- |
| .alpha.-Monostearin |
| Monoglyceryl stearate |
| Glycerol alpha-monostearate |
| Cefatin |
| Dermagine |
| Monelgin |
| Sedetine |
| Admul |
| Orbon |
| Citomulgan M |
| 2,3-Dihydroxypropyl stearate |
| Drewmulse V |
| Cerasynt S |
| Drewmulse TP |
| Tegin 515 |
| Cerasynt SE |
| Cerasynt WM |
| Cyclochem GMS |
| Drumulse AA |
| Protachem GMS |
| Witconol MS |
| Witconol MST |
| FEMA No. 2527 |
| Glyceryl stearates |
| Monostearate (glyceride) |
| Unimate GMS |
| Glyceryl monooctadecanoate |
| Ogeen M |
| Emcol CA |
| Emcol MSK |
| Hodag GMS |
| Ogeen GRB |
| Ogeen MAV |
| Aldo MS |
| Aldo HMS |
| Armostat 801 |
| Kessco 40 |
| Stearic monoglyceride |
| Abracol S.L.G. |
| Arlacel 161 |
| Arlacel 169 |
| Imwitor 191 |
| Imwitor 900K |
| NSC 3875 |
| Atmul 67 |
| Atmul 84 |
| Starfol GMS 450 |
| Starfol GMS 600 |
| Starfol GMS 900 |
| Cerasynt 1000-D |
| Emerest 2401 |
| Aldo-28 |
| Aldo-72 |
| Atmos 150 |
| Atmul 124 |
| Estol 603 |
| Ogeen 515 |
| Tegin 503 |
| Grocor 5500 |
| Grocor 6000 |
| Glycerol stearate, pure |
| Stearic acid alpha-monoglyceride |
| Cremophor gmsk |
| Glyceryl 1-octadecanoate |
| Cerasynt-sd |
| Lonzest gms |
| Cutina gms |
| Lipo GMS 410 |
| Lipo GMS 450 |
| Lipo GMS 600 |
| glycerol stearate |
| 1-MONOSTEAROYL-rac-GLYCEROL |
| Nikkol mgs-a |
| Glyceryl monopalmitostearate |
| USAF KE-7 |
| 1-octadecanoyl-rac-glycerol |
| EMUL P.7 |
| EINECS 204-664-4 |
| EINECS 245-121-1 |
| UNII-230OU9XXE4 |
| Celinhol - A |
| Stearic acid, monoester with glycerol |
| Glycerol .alpha.-monostearate |
| Glyceroli monostearas |
| Myvaplex 600 |
| Glycerol monostearate, purified |
| Imwitor 491 |
| Sorbon mg-100 |
| 22610-63-5 |
| Cithrol gms 0400 |
| UNII-258491E1RZ |
| NSC3875 |
| Stearic acid .alpha.-monoglyceride |
| (1)-2,3-Dihydroxypropyl stearate |
| MONOSTEARIN (L) |
| C21H42O4 |
| NSC-3875 |
| 1-Monooctadecanoylglycerol |
| EINECS 250-705-4 |
| 1,2,3-Propanetriol monooctadecanoate |
| Octadecanoic acid, ester with 1,2,3-propanetriol |
| GLYCERYL 1-STEARATE |
| TEGIN 90 |
| 1-O-Octadecanoyl-2n-glycerol |
| AI3-00966 |
| MG(18:0/0:0/0:0)[rac] |
| 230OU9XXE4 |
| DTXSID7029160 |
| Glyceryl monostearate [JAN:NF] |
| CHEBI:75555 |
| EC 250-705-4 |
| GLYCERYL MONOSTEARATE 40-50 |
| Octadecanoic acid, monoester with 1,2,3-propanetriol |
| 258491E1RZ |
| 1-Stearoyl-rac-glycerol (90per cent) |
| 83138-62-9 |
| 85666-92-8 |
| NCGC00164529-01 |
| (+/-)-2,3-DIHYDROXYPROPYL OCTADECANOATE |
| DTXCID909160 |
| Octadecanoic acid, 2,3-dihydroxypropyl ester, (A+/-)- |
| MFCD00036186 |
| CAS-123-94-4 |
| rac-Glycerol 1-stearate |
| EINECS 238-880-5 |
| 1-Monooctadecanoyl-rac-glycerol |
| Celinhol-A |
| MG 18:0 |
| (+/-)-2,3-Dihydroxypropyl octadecanoate; 1-Glyceryl stearate; 1-Monooctadecanoylglycerol; 1-Monostearin |
| Eastman 600 |
| 1-O-stearoylglycerol |
| 1-octadecanoylglycerol |
| Glycerol Mono Stearate |
| rac-octadecanoylglycerol |
| glycerol 1-octadecanoate |
| rac-glyceryl monostearate |
| Glycerol .alpha.-sterate |
| rac-1-monostearoylglycerol |
| DSSTox_CID_9160 |
| Dur-Em 117 |
| Monoglycerides, c16-18 |
| (+-)-1-stearoylglycerol |
| SCHEMBL4488 |
| (+-)-glyceryl monostearate |
| Geleol mono and diglycerides |
| DSSTox_RID_78757 |
| DSSTox_GSID_29304 |
| Glycerol monostearate (GMS) |
| (+-)-1-monostearoylglycerol |
| (+-)-1-octadecanoylglycerol |
| Glycerides, C16-18 mono- |
| Glycerol monostearate 40-55 |
| GLYCERYL STEARATE (II) |
| CHEMBL255696 |
| 2,3-Dihydroxypropyl stearate # |
| DTXSID7027968 |
| CHEBI:75557 |
| 1-Stearoyl-rac-glycerol (90%) |
| GLYCERYL MONOSTEARATE (II) |
| Glyceryl monostearate (JP17/NF) |
| 1-Stearoyl-rac-glycerol, >=99% |
| MAG 18:0 |
| (+-)-2,3-dihydroxypropyl stearate |
| EINECS 293-208-8 |
| Tox21_112160 |
| Tox21_202573 |
| Tox21_301104 |
| LMGL01010003 |
| rac-2,3-dihydroxypropyl octadecanoate |
| AKOS015901589 |
| STEARATE, 2,3-DIHYDROXYPROPYL |
| Tox21_112160_1 |
| DB11250 |
| (+-)-2,3-dihydroxypropyl octadecanoate |
| NCGC00164529-02 |
| NCGC00164529-03 |
| NCGC00164529-04 |
| NCGC00255004-01 |
| NCGC00260122-01 |
| Octadecanoic acid,3-dihydroxypropyl ester |
| 1,2,3-Propanetriol 1-octadecanoyl ester |
| BS-50505 |
| STEARATE, 2,3-DIHYDROXYPROP-1-YL |
| C18-H36-O2.(C3H8-O3)x- |
| CAS-11099-07-3 |
| LS-164168 |
| FT-0626740 |
| FT-0626748 |
| FT-0674656 |
| G0085 |
| Octadecanoic acid, 2.3-dihydroxypropyl ester |
| C18-H36-O2.x-(C3-H8-O3)x- |
| D01947 |
| EC 293-208-8 |
| F71433 |
| S-7950 |
| 500-265-7 (NLP #) |
| A890632 |
| A903419 |
| SR-01000944874 |
| Octadecanoic acid monoester with 1,2,3-propanetriol |
| Octadecanoic acid, monester with 1,2,3-propanetriol |
| Q-201168 |
| Q5572563 |
| SR-01000944874-1 |
| W-110285 |
| Glyceryl monostearate; (Octadecanoic acid, monoester with 1,2,3-propanetriol) |
| ()-2,3-Dihydroxypropyl octadecanoate; 1-Glyceryl stearate; 1-Monooctadecanoylglycerol; 1-Monostearin |
| 342394-34-7 |
| 37349-34-1 |
| 8029-22-9 |
| InChI=1/C21H42O4/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-21(24)25-19-20(23)18-22/h20,22-23H,2-19H2,1H |
| Octadecanoic acid, monoester with 3,3'-((2-hydroxy-1,3-propanediyl)bis(oxy))bis(1,2-propanediol- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|