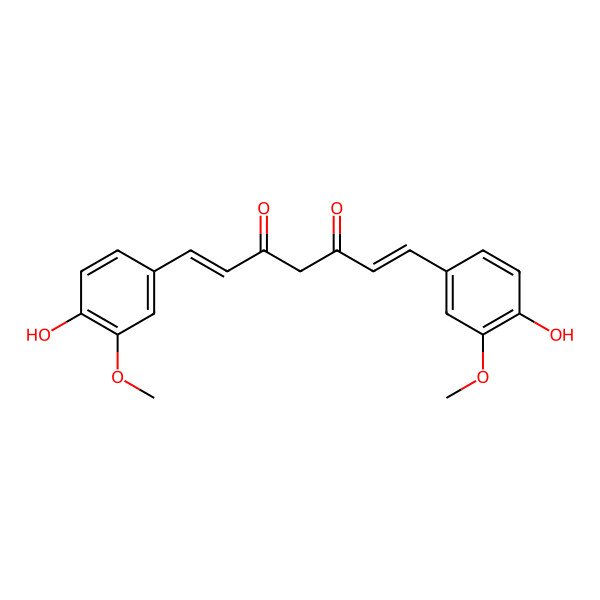
Curcumin
| Internal ID | 561884c3-72ef-48fb-9020-f2b5254fffbd |
| Taxonomy | Phenylpropanoids and polyketides > Diarylheptanoids > Linear diarylheptanoids > Curcuminoids |
| IUPAC Name | (1E,6E)-1,7-bis(4-hydroxy-3-methoxyphenyl)hepta-1,6-diene-3,5-dione |
| SMILES (Canonical) | COC1=C(C=CC(=C1)C=CC(=O)CC(=O)C=CC2=CC(=C(C=C2)O)OC)O |
| SMILES (Isomeric) | COC1=C(C=CC(=C1)/C=C/C(=O)CC(=O)/C=C/C2=CC(=C(C=C2)O)OC)O |
| InChI | InChI=1S/C21H20O6/c1-26-20-11-14(5-9-18(20)24)3-7-16(22)13-17(23)8-4-15-6-10-19(25)21(12-15)27-2/h3-12,24-25H,13H2,1-2H3/b7-3+,8-4+ |
| InChI Key | VFLDPWHFBUODDF-FCXRPNKRSA-N |
| Popularity | 30,118 references in papers |
| Molecular Formula | C21H20O6 |
| Molecular Weight | 368.40 g/mol |
| Exact Mass | 368.12598835 g/mol |
| Topological Polar Surface Area (TPSA) | 93.10 Ų |
| XlogP | 3.20 |
| 458-37-7 |
| Diferuloylmethane |
| Natural yellow 3 |
| Indian saffron |
| Kacha haldi |
| Curcuma |
| Gelbwurz |
| Haldar |
| Curcumin I |
| Souchet |
| There are more than 10 synonyms. If you wish to see them all click here. |
| Target | Value | Probability (raw) | Probability (%) |
|---|---|---|---|
| No predicted properties yet! | |||
Proven Targets:
| CHEMBL ID | UniProt ID | Name | Min activity | Assay type | Source |
|---|---|---|---|---|---|
| CHEMBL1293255 | P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD+] |
39810.7 nM 14125.4 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL3577 | P00352 | Aldehyde dehydrogenase 1A1 |
28183.8 nM 8912.5 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL3687 | P18054 | Arachidonate 12-lipoxygenase |
19952.6 nM |
Potency |
via CMAUP
|
| CHEMBL2903 | P16050 | Arachidonate 15-lipoxygenase |
15848.9 nM 12589.3 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL2487 | P05067 | Beta amyloid A4 protein |
11300 nM 3100 nM 1500 nM 21810 nM 630 nM |
IC50 IC50 IC50 IC50 IC50 |
PMID: 26783181
PMID: 27190601 PMID: 27190601 PMID: 27353888 via Super-PRED |
| CHEMBL5990 | P38398 | Breast cancer type 1 susceptibility protein |
5011.9 nM 5011.9 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL4801 | P29466 | Caspase-1 |
31622.8 nM 15848.9 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL3468 | P55210 | Caspase-7 |
12589.3 nM |
Potency |
via CMAUP
|
| CHEMBL4096 | P04637 | Cellular tumor antigen p53 |
39810.7 nM 39810.7 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL4081 | P13726 | Coagulation factor III |
13.16 nM |
IC50 |
via Super-PRED
|
| CHEMBL5747 | Q92793 | CREB-binding protein |
25000 nM |
IC50 |
PMID: 26701186
|
| CHEMBL340 | P08684 | Cytochrome P450 3A4 |
15848.9 nM 12589.3 nM 15848.9 nM 12589.3 nM |
Potency Potency Potency Potency |
via CMAUP
via CMAUP via CMAUP via CMAUP |
| CHEMBL2392 | P06746 | DNA polymerase beta |
44668.4 nM |
Potency |
via CMAUP
|
| CHEMBL4159 | Q99714 | Endoplasmic reticulum-associated amyloid beta-peptide-binding protein |
31622.8 nM 3981.1 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL262 | P49841 | Glycogen synthase kinase-3 beta |
17950 nM |
IC50 |
PMID: 26696252
|
| CHEMBL2424 | Q04760 | Glyoxalase I |
5 nM |
Ki |
via Super-PRED
|
| CHEMBL4331 | P68871 | Hemoglobin beta chain |
39810.7 nM |
Potency |
via CMAUP
|
| CHEMBL3784 | Q09472 | Histone acetyltransferase p300 |
25000 nM 6500 nM |
IC50 IC50 |
PMID: 26701186
PMID: 26701186 |
| CHEMBL1293299 | Q03164 | Histone-lysine N-methyltransferase MLL |
14125.4 nM |
Potency |
via CMAUP
|
| CHEMBL5514 | P42858 | Huntingtin |
3548.1 nM |
Potency |
via CMAUP
|
| CHEMBL4261 | Q16665 | Hypoxia-inducible factor 1 alpha |
6309.6 nM 6309.6 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL1293226 | B2RXH2 | Lysine-specific demethylase 4D-like |
35481.3 nM 35481.3 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL2608 | P10253 | Lysosomal alpha-glucosidase |
39506.8 nM |
Potency |
via CMAUP
|
| CHEMBL4040 | P28482 | MAP kinase ERK2 |
39810.7 nM 31622.8 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL1293224 | P10636 | Microtubule-associated protein tau |
3548.1 nM 3548.1 nM 12589.3 nM |
Potency Potency Potency |
via CMAUP
via CMAUP via CMAUP |
| CHEMBL1951 | P21397 | Monoamine oxidase A |
710 nM 710 nM |
Ki Ki |
PMID: 26819666
via Super-PRED |
| CHEMBL2039 | P27338 | Monoamine oxidase B |
21500 nM |
Ki |
PMID: 26819666
|
| CHEMBL5162 | Q6W5P4 | Neuropeptide S receptor |
316.2 nM 316.2 nM |
Potency Potency |
via CMAUP
via Super-PRED |
| CHEMBL1293235 | P02545 | Prelamin-A/C |
1778.3 nM 35481.3 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL5658 | O14684 | Prostaglandin E synthase |
200 nM 200 nM 220 nM |
IC50 IC50 IC50 |
via Super-PRED
DOI: 10.1039/C5MD00278H PMID: 26588603 |
| CHEMBL1075189 | P14618 | Pyruvate kinase isozymes M1/M2 |
28183.8 nM 28183.8 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL1963 | P16473 | Thyroid stimulating hormone receptor |
10000 nM 10000 nM |
Potency Potency |
via CMAUP
via CMAUP |
| CHEMBL5804 | Q9NR96 | Toll-like receptor 9 |
8362 nM |
IC50 |
via CMAUP
|
| CHEMBL1075138 | Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 |
1122 nM 2511.9 nM |
Potency Potency |
via CMAUP
via CMAUP |
Predicted Targets (via Super-PRED):
| CHEMBL ID | UniProt ID | Name | Probability | Model accuracy |
|---|---|---|---|---|
| CHEMBL5619 | P27695 | DNA-(apurinic or apyrimidinic site) lyase | 98.55% | 91.11% |
| CHEMBL1075094 | Q16236 | Nuclear factor erythroid 2-related factor 2 | 95.53% | 96.00% |
| CHEMBL3251 | P19838 | Nuclear factor NF-kappa-B p105 subunit | 94.05% | 96.09% |
| CHEMBL1293249 | Q13887 | Kruppel-like factor 5 | 94.04% | 86.33% |
| CHEMBL3108638 | O15164 | Transcription intermediary factor 1-alpha | 91.85% | 95.56% |
| CHEMBL3060 | Q9Y345 | Glycine transporter 2 | 90.91% | 99.17% |
| CHEMBL4208 | P20618 | Proteasome component C5 | 88.74% | 90.00% |
| CHEMBL2535 | P11166 | Glucose transporter | 87.48% | 98.75% |
| CHEMBL3194 | P02766 | Transthyretin | 87.16% | 90.71% |
| CHEMBL225 | P28335 | Serotonin 2c (5-HT2c) receptor | 86.97% | 89.62% |
| CHEMBL1806 | P11388 | DNA topoisomerase II alpha | 84.81% | 89.00% |
| CHEMBL4203 | Q9HAZ1 | Dual specificity protein kinase CLK4 | 84.69% | 94.45% |
| CHEMBL3401 | O75469 | Pregnane X receptor | 80.73% | 94.73% |
Below are displayed all the plants proven (via scientific papers) to contain this
compound!
To see more specific details click the taxa you are interested in.
To see more specific details click the taxa you are interested in.