| 1107-26-2 |
| 8'-Apo-beta-carotenal |
| beta-apo-8'-carotenal |
| C.I. Food Orange 6 |
| 8'-Apo-beta-caroten-8'-al |
| beta-apo-Carotenal |
| C Orange 16 |
| beta-Apocarotenal |
| Food orange 6 |
| 8'Apo-beta,psi-carotenal |
| trans-beta-Apo-8'-carotenal |
| beta-Apo-8'-carotenal (C30) |
| all-trans-beta-Apo-8'-carotenal |
| BETA-APO-8'-CAROTENAL (TRANS) |
| CCRIS 7933 |
| all-trans-8'-apo-beta-carotenal |
| E160E |
| E 160 e |
| 8'-Apo-beta-carotenal, all-trans- |
| EINECS 214-171-6 |
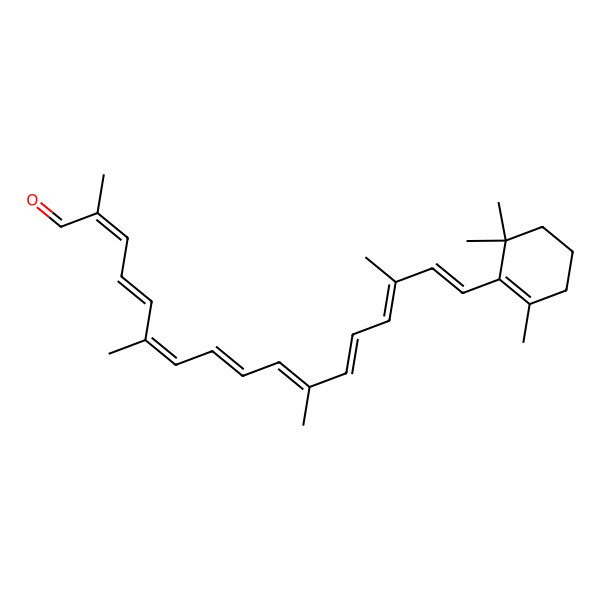
| (2E,4E,6E,8E,10E,12E,14E,16E)-2,6,11,15-tetramethyl-17-(2,6,6-trimethylcyclohexen-1-yl)heptadeca-2,4,6,8,10,12,14,16-octaenal |
| .beta.-apo-8'-Carotenal |
| 8'-Apo-beta,psi-carotenal |
| BRN 2064131 |
| CI 40820 |
| UNII-V22N3E2U32 |
| Beta-apo-8'-carotenal(trans) |
| C.I. 40820 |
| 8'-apo-beta,psi-caroten-8'-al |
| V22N3E2U32 |
| 2,4,6,8,10,12,14,16-Heptadecaoctaenal, 2,6,11,15-tetramethyl-17-(2,6,6-trimethyl-1-cyclohexen-1-yl)-, (2E,4E,6E,8E,10E,12E,14E,16E)- |
| 8'-APO-.BETA.,.PSI.-CAROTENAL |
| 2,4,6,8,10,12,14,16-Heptadecaoctaenal, 2,6,11,15-tetramethyl-17-(2,6,6-trimethyl-1-cyclohexen-1-yl)-, (all-E)- |
| 4-07-00-01782 (Beilstein Handbook Reference) |
| 8'-Apoaldehyde |
| -po-8'-arotenal |
| CI Food Orange 6 |
| |A-Apo-8'-carotenal |
| trans-b-apo-8'-carotenal |
| trans-?-Apo-8?-carotenal |
| APOCAROTENAL [MART.] |
| Color Index No. 40820 |
| Color Index Number 40820 |
| Colour Index No. 40820 |
| APOCAROTENAL [USP-RS] |
| Colour Index Number 40820 |
| INS NO.160E |
| SCHEMBL341809 |
| INS-160E |
| SCHEMBL2768511 |
| trans- beta -Apo-8'-carotenal |
| CHEBI:53154 |
| C30H40O |
| DTXSID20883251 |
| DFMMVLFMMAQXHZ-DOKBYWHISA-N |
| 8'-APO-.BETA.-CAROTENAL |
| CAROTENAL, BETA-APO-8'- |
| CI 40820 [INCI] |
| HY-N6677 |
| 8'-Apo-.beta.,.pseudo.-carotenal |
| BETA-APO-8-CAROTENAL(TRANS) |
| C30-H40-O |
| E-160E |
| NSC374897 |
| AKOS024386366 |
| BETA-APO-8'-CAROTENAL [FCC] |
| 8'-Apo-.beta.-carotenal, all-trans- |
| NSC-374897 |
| NCGC00187588-01 |
| LS-21455 |
| XA167402 |
| CI(1975)NO.40820 |
| CS-0099621 |
| S5846 |
| CAROTENAL, 8'-APO-.BETA., .PSI.- |
| C19728 |
| F82418 |
| trans-beta-Apo-8'-carotenal, >=96.0% (UV) |
| Q274972 |
| J-002468 |
| Z2583134452 |
| Apocarotenal, United States Pharmacopeia (USP) Reference Standard |
| (2E,4E,6E,8E,10E,12E,14E)-2,6,11,15-Tetramethyl-17-(2,6,6-trimethyl-1-cyclohexenyl)-2,4,6,8,10,12,14,16-heptadecaoctaenal |
| (2E,4E,6E,8E,10E,12E,14E,16E)-2,6,11,15-tetramethyl-17-(2,6,6-trimethylcyclohex-1-enyl)heptadeca-2,4,6,8,10,12,14,16-octaenal |
|
There are more than 10 synonyms. If you wish to see them all click here.
|