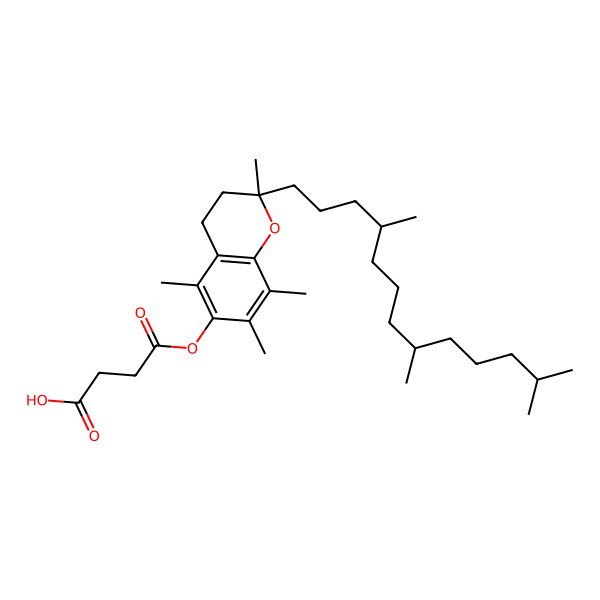
| 4345-03-3 |
| alpha-Tocopheryl succinate |
| D-alpha-Tocopherol succinate |
| alpha-tocopherol succinate |
| tocopherol succinate |
| D-ALPHA-TOCOPHERYL SUCCINATE |
| Vitamin E hemisuccinate |
| Vitamine E succinate |
| Alpha-tocopherol succinate, d- |
| d-alpha-Tocopherol acid succinate |
| D-|A-Tocopherol succinate |
| alpha tocopheryl acid succinate |
| alpha-Tocopheryl acid succinate |
| (+)-alpha-Tocopheryl succinate |
| alpha-Vitamin E succinate |
| LU4B53JYVE |
| alpha-Tocopherol, succinate |
| NSC 173849 |
| .alpha.-Tocopherol succinate |
| alpha-Tocopherol hemisuccinate |
| CCRIS 4734 |
| alpha-Tocopheryl succinate, D- |
| 4-oxo-4-[[(2R)-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-3,4-dihydrochromen-6-yl]oxy]butanoic acid |
| DTXSID2026151 |
| D-alpha tocopheryl acid succinate |
| d-alpha-Tocopheryl acid succinate |
| Tocopherol acid succinate, alpha- |
| alpha-Tocopheryl hydrogen succinate |
| alpha-Tocopherol acid succinate, D- |
| EINECS 224-403-8 |
| EINECS 241-433-7 |
| 4-Oxo-4-(((R)-2,5,7,8-tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)chroman-6-yl)oxy)butanoic acid |
| tocopheryl hemisuccinate |
| NCGC00167561-01 |
| TOCOPHERYL ACID SUCCINATE,D-ALPHA |
| .ALPHA.-TOCOPHEROL SUCCINATE, D- |
| tocopheryl acid succinate |
| DTXCID706151 |
| |A-Tocopheryl Succinate |
| C33H54O5 |
| 2,5,7,8-Tetramethyl-2-(4,8,12-trimethyltridecyl)-6-chromanyl hydrogen succinate, (+)- |
| 6-Chromanol, 2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-, hydrogen succinate, (+)- |
| Mono(2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-6-chromanyl) succinate, (+)- |
| 4-oxo-4-{[(2R)-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-3,4-dihydro-2H-1-benzopyran-6-yl]oxy}butanoic acid |
| Butanedioic acid, 1-((2R)-3,4-dihydro-2,5,7,8-tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl) ester |
| Butanedioic acid, mono((2R)-3,4-dihydro-2,5,7,8-tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl) ester |
| Butanedioic acid, mono(3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl) ester, (2R-(2R*(4R*,8R*)))- |
| Mono(3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl) butanedioate, (2R-(2R*(4R*,8R*)))- |
| Succinic acid, mono(2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-6-chromanyl) ester, (+)- |
| CAS-4345-03-3 |
| Vitamin E acid succinate |
| Covitol 1210 |
| D-.alpha.-Tocopherol succinate |
| D-.alpha.-Tocopheryl succinate |
| NSC173849 |
| C33-H54-O5 |
| NSC-173849 |
| Vitamin-E Dragees |
| (3,4-Dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl) hydrogen succinate |
| [3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl] hydrogen succinate |
| 4-oxo-4-((R)-2,5,7,8-tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)chroman-6-yloxy)butanoic acid |
| Butanedioic acid, mono[(2R)-3,4-dihydro-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-2H-1-benzopyran-6-yl] ester |
| Tocopheryl succinate |
| Dal-E |
| CV 104 |
| MFCD00072055 |
| -Tocopheryl Succinate |
| UNII-LU4B53JYVE |
| D-alpha-Tocopherolsuccinate |
| D-alpha-tocopheryl-succinate |
| .alpha.-tocopheryl succinate |
| White-e [veterinary] (TN) |
| CHEMBL81421 |
| SCHEMBL134422 |
| D- alpha -Tocopheryl Succinate |
| alpha-Tocopherol, succinate, D- |
| .alpha.-tocopheryl succinate, d- |
| CHEBI:135821 |
| Tocopheryl Acid Succinate, d-Alpha |
| TOCOPHERYL SUCCINATE [INCI] |
| Butanedioic acid, mono(3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl) ester |
| Butanedioic acid, mono[3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl] ester |
| Tox21_112556 |
| Tox21_200568 |
| BDBM50458511 |
| D-alpha-Tocopheryl hydrogen succinate |
| LS-120 |
| AKOS015902063 |
| Dal-Vita brand of vitamin E succinate |
| Wiedemann brand of vitamin E succinate |
| (+)-.alpha.-Tocopherol acid succinate |
| AC-1132 |
| CCG-207945 |
| DB14001 |
| RRR-alpha-Tocopheryl Hydrogen Succinate |
| NCGC00167561-02 |
| NCGC00167561-03 |
| NCGC00258122-01 |
| TOCOPHERYL ACID SUCCINATE [VANDF] |
| 3-{[(2R)-2-((4R,8R)-4,8,12-trimethyltridecyl)-2,5,7,8-tetramethylchroman-6-yl] oxycarbonyl}propanoic acid |
| 4-oxo-4-[(2R)-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]chroman-6-yl]oxy-butanoic acid |
| AS-75106 |
| Butanedioic acid,mono[(2R)-3,4-dihydro-2,5,7,8-tetramethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-2H-1-benzopyran-6-yl] ester |
| J24.824J |
| .ALPHA.-TOCOPHEROL SUCCINATE [MI] |
| DL-ALPHA-TOCOPHEROLHYDROGENSUCCINATE |
| RRR-ALPHA-TOCOPHERYL ACID SUCCINATE |
| TOCOPHERYL ACID SUCCINATE [WHO-DD] |
| HY-131553 |
| CS-0136520 |
| T2628 |
| VITAMIN E (ALPHA-TOCOPHERYL SUCCINATE) |
| C90299 |
| D08612 |
| D-ALPHA TOCOFERIL ACID SUCCINATE [MART.] |
| D-alpha-Tocopherol succinate, analytical standard |
| RRR-.ALPHA.-TOCOPHERYL HYDROGEN SUCCINATE |
| C033716 |
| EN300-23011947 |
| RRR-ALPHA-TOCOPHERYL ACID SUCCINATE [FCC] |
| SR-01000883728 |
| TOCOPHERYL ACID SUCCINATE,D-ALPHA [VANDF] |
| SR-01000883728-1 |
| Q27283185 |
| D-alpha-Tocopherol succinate, semisynthetic, 1210 IU/g |
| Z2216889767 |
| D-alpha-Tocopherol succinate, BioXtra, >=98.0% (HPLC) |
| RRR-.ALPHA.-TOCOPHERYL HYDROGEN SUCCINATE [EP IMPURITY] |
| Alpha tocopheryl acid succinate, United States Pharmacopeia (USP) Reference Standard |
| RRR-alpha-Tocopheryl hydrogen succinate, European Pharmacopoeia (EP) Reference Standard |
| 4-Oxo-4-(((R)-2,5,7,8-tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)chroman-6-yl)oxy)butanoicacid |
| 55134-51-5 |
| Butanedioic acid, mono(3,4-dihydro-2,5,7,8-tetramethyl-2-(4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl) ester, (2R-(2R*( |
| mono((2R)-3,4-dihydro-2,5,7,8-tetramethyl-2-((4R,8R)-4,8,12-trimethyltridecyl)-2H-1-benzopyran-6-yl) butanedioate |
| Tocopheryl Acid Succinate, a, Pharmaceutical Secondary Standard; Certified Reference Material |
|
There are more than 10 synonyms. If you wish to see them all click here.
|