| 97-30-3 |
| alpha-Methylglucoside |
| Methyl-alpha-D-glucopyranoside |
| Methyl alpha-D-glucoside |
| alpha-Methyl-D-glucoside |
| alpha-D-Methylglucoside |
| alpha-Methyl D-glucose ether |
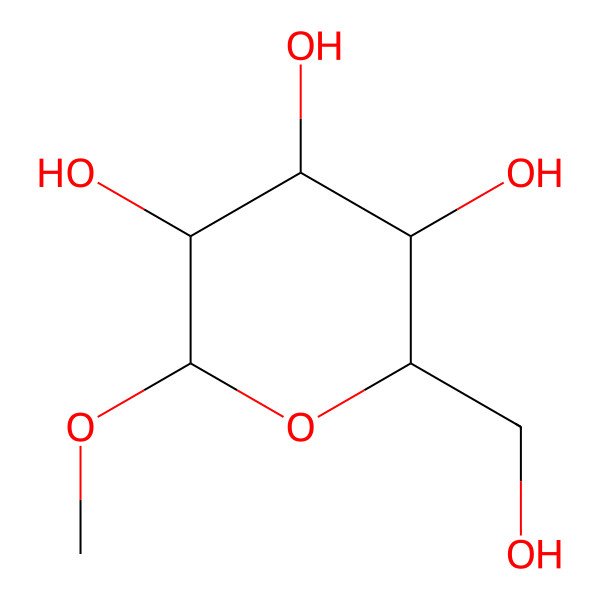
| (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-methoxyoxane-3,4,5-triol |
| alpha-MDG |
| Me alpha-Glc |
| .alpha.-D-Glucopyranoside, methyl |
| alpha-D-methyl glucoside |
| 1-O-methyl-alpha-D-glucoside |
| (2R,3S,4S,5R,6S)-2-(Hydroxymethyl)-6-methoxytetrahydro-2H-pyran-3,4,5-triol |
| Methyl alpha-D-glucoside (VAN) |
| 1-O-methyl-alpha-D-glucopyranose |
| CHEBI:320061 |
| .alpha.-Methylglucoside |
| 1-O-methyl-alpha-D-glucopyranoside |
| EINECS 202-571-3 |
| alpha-D-Glucoside, methyl |
| UNII-QCF122NF3R |
| NSC 102101 |
| Methyl .alpha.-D-glucoside |
| QCF122NF3R |
| AI3-18790 |
| alpha-D-Glucopyranoside, methyl |
| .alpha.-Methyl D-glucose ether |
| DTXSID7026605 |
| methyl a-d-glucopyranoside |
| alpha-methyl-D-glucose pyranoside |
| Methyl .alpha.-D-glucopyranoside |
| Methyl ?-D-Glucopyranoside |
| Glucopyranoside, methyl, alpha-D- |
| C7H14O6 |
| a-methylglucoside |
| GYP |
| METHYLGLUCOSIDE, ALPHA |
| methyl glucose |
| alphaMG |
| MalphaDG |
| Glucopyranoside, methyl, .alpha.-D- |
| Me-alpha-Glc |
| NSC-102101 |
| NSC-214092 |
| MFCD00064086 |
| a-methyl-D-glucoside |
| 1ws4 |
| Methyl hexopyranoside # |
| a-D-methyl glucopyranoside |
| D0A3FF |
| Epitope ID:150073 |
| methyl alpha-D-glucopyranose |
| SCHEMBL50473 |
| ALPHA-METHYL GLUCOSIDE |
| .alpha.-D-Glucoside, methyl |
| alpha-D-methyl glucopyranoside |
| .alpha.-Methyl-(d)-glucoside |
| .alpha.-d-Methylglucopyranoside |
| CHEMBL131853 |
| DTXCID806605 |
| GTPL4640 |
| Methyl alpha -D-glucopyranoside |
| Glucoside, methyl, .alpha.-D- |
| BDBM20876 |
| methyl alpha-(D)-glucopyranoside |
| Tox21_200430 |
| [14C]alpha-methyl-D-glucopyranoside |
| .ALPHA.-METHYLGLUCOSIDE [MI] |
| alpha-methyl-D-glucopyranoside (AMG) |
| AKOS015896548 |
| CS-W021034 |
| CAS-97-30-3 |
| Methyl alpha-D-glucopyranoside, >=99% |
| 1-O-METHYL-.ALPHA.-D-GLUCOSIDE |
| METHYL .ALPHA.-D-(+)-GLUCOSIDE |
| NCGC00257984-01 |
| AS-14322 |
| LS-180668 |
| M0228 |
| 1-O-METHYL-.ALPHA.-D-GLUCOPYRANOSIDE |
| EN300-92952 |
| P16421 |
| ethyl1-methyl-3-phenyl-1H-pyrazole-5-carboxylate |
| Q27074402 |
| BE2A51F8-C121-4E44-89AB-05A013F8AA57 |
| Methyl alpha-D-glucopyranoside, for microbiology, >=99.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|