Gefitinib
| Internal ID | 3de4c890-91bd-4a52-9fb2-455163def98d |
| Taxonomy | Organoheterocyclic compounds > Diazanaphthalenes > Benzodiazines > Quinazolines > Quinazolinamines |
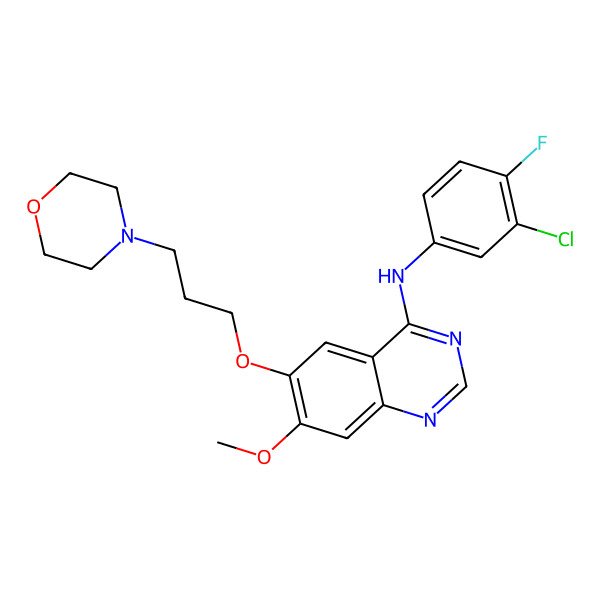
| IUPAC Name | N-(3-chloro-4-fluorophenyl)-7-methoxy-6-(3-morpholin-4-ylpropoxy)quinazolin-4-amine |
| SMILES (Canonical) | COC1=C(C=C2C(=C1)N=CN=C2NC3=CC(=C(C=C3)F)Cl)OCCCN4CCOCC4 |
| SMILES (Isomeric) | COC1=C(C=C2C(=C1)N=CN=C2NC3=CC(=C(C=C3)F)Cl)OCCCN4CCOCC4 |
| InChI | InChI=1S/C22H24ClFN4O3/c1-29-20-13-19-16(12-21(20)31-8-2-5-28-6-9-30-10-7-28)22(26-14-25-19)27-15-3-4-18(24)17(23)11-15/h3-4,11-14H,2,5-10H2,1H3,(H,25,26,27) |
| InChI Key | XGALLCVXEZPNRQ-UHFFFAOYSA-N |
| Popularity | 16,381 references in papers |
| Molecular Formula | C22H24ClFN4O3 |
| Molecular Weight | 446.90 g/mol |
| Exact Mass | 446.1520965 g/mol |
| Topological Polar Surface Area (TPSA) | 68.70 Ų |
| XlogP | 4.10 |
| Atomic LogP (AlogP) | 4.28 |
| H-Bond Acceptor | 7 |
| H-Bond Donor | 1 |
| Rotatable Bonds | 8 |
| 184475-35-2 |
| Iressa |
| ZD1839 |
| N-(3-Chloro-4-fluorophenyl)-7-methoxy-6-(3-morpholinopropoxy)quinazolin-4-amine |
| Irressat |
| gefitinib (zd1839) |
| ZD 1839 |
| ZD-1839 |
| Gefitinib [USAN] |
| N-(3-chloro-4-fluorophenyl)-7-methoxy-6-[3-(morpholin-4-yl)propoxy]quinazolin-4-amine |
| There are more than 10 synonyms. If you wish to see them all click here. |
| Target | Value | Probability (raw) | Probability (%) |
|---|---|---|---|
| Human Intestinal Absorption | + | 0.9951 | 99.51% |
| Caco-2 | - | 0.5623 | 56.23% |
| Blood Brain Barrier | + | 0.8000 | 80.00% |
| Human oral bioavailability | + | 0.9000 | 90.00% |
| Subcellular localzation | Lysosomes | 0.4920 | 49.20% |
| OATP2B1 inhibitior | - | 0.8460 | 84.60% |
| OATP1B1 inhibitior | + | 0.9220 | 92.20% |
| OATP1B3 inhibitior | + | 0.9455 | 94.55% |
| MATE1 inhibitior | - | 0.9000 | 90.00% |
| OCT2 inhibitior | - | 0.5750 | 57.50% |
| BSEP inhibitior | + | 0.9938 | 99.38% |
| P-glycoprotein inhibitior | + | 0.8836 | 88.36% |
| P-glycoprotein substrate | - | 0.8125 | 81.25% |
| CYP3A4 substrate | + | 0.6774 | 67.74% |
| CYP2C9 substrate | - | 0.8176 | 81.76% |
| CYP2D6 substrate | + | 0.4132 | 41.32% |
| CYP3A4 inhibition | + | 0.8206 | 82.06% |
| CYP2C9 inhibition | - | 0.6272 | 62.72% |
| CYP2C19 inhibition | + | 0.6440 | 64.40% |
| CYP2D6 inhibition | - | 0.6536 | 65.36% |
| CYP1A2 inhibition | + | 0.7516 | 75.16% |
| CYP2C8 inhibition | + | 0.8738 | 87.38% |
| CYP inhibitory promiscuity | + | 0.9309 | 93.09% |
| UGT catelyzed | - | 0.0000 | 0.00% |
| Carcinogenicity (binary) | - | 0.8318 | 83.18% |
| Carcinogenicity (trinary) | Non-required | 0.5807 | 58.07% |
| Eye corrosion | - | 0.9886 | 98.86% |
| Eye irritation | - | 0.9737 | 97.37% |
| Skin irritation | - | 0.7733 | 77.33% |
| Skin corrosion | - | 0.9467 | 94.67% |
| Ames mutagenesis | + | 0.5700 | 57.00% |
| Human Ether-a-go-go-Related Gene inhibition | + | 0.8964 | 89.64% |
| Micronuclear | + | 0.8900 | 89.00% |
| Hepatotoxicity | + | 0.8125 | 81.25% |
| skin sensitisation | - | 0.8475 | 84.75% |
| Respiratory toxicity | + | 0.9778 | 97.78% |
| Reproductive toxicity | + | 0.9333 | 93.33% |
| Mitochondrial toxicity | + | 1.0000 | 100.00% |
| Nephrotoxicity | - | 0.8449 | 84.49% |
| Acute Oral Toxicity (c) | III | 0.7006 | 70.06% |
| Estrogen receptor binding | + | 0.8490 | 84.90% |
| Androgen receptor binding | + | 0.6230 | 62.30% |
| Thyroid receptor binding | + | 0.7370 | 73.70% |
| Glucocorticoid receptor binding | + | 0.6941 | 69.41% |
| Aromatase binding | + | 0.7985 | 79.85% |
| PPAR gamma | + | 0.6857 | 68.57% |
| Honey bee toxicity | - | 0.9081 | 90.81% |
| Biodegradation | - | 0.9000 | 90.00% |
| Crustacea aquatic toxicity | + | 0.6800 | 68.00% |
| Fish aquatic toxicity | - | 0.4206 | 42.06% |
Proven Targets:
| CHEMBL ID | UniProt ID | Name | Min activity | Assay type | Source |
|---|---|---|---|---|---|
| CHEMBL5393 | Q9UNQ0 | ATP-binding cassette sub-family G member 2 |
400 nM 1100 nM 1260 nM |
IC50 IC50 IC50 |
PMID: 15155841
PMID: 19932960 PMID: 19932960 |
| CHEMBL4822 | P56817 | Beta-secretase 1 |
20000 nM |
IC50 |
PMID: 27046190
|
| CHEMBL5650 | Q8TDC3 | BR serine/threonine-protein kinase 1 |
3162.28 nM 3162.28 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL4179 | P45984 | c-Jun N-terminal kinase 2 |
2511.89 nM 2511.89 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL2637 | P53779 | c-Jun N-terminal kinase 3 |
794.33 nM 794.33 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL2801 | Q13557 | CaM kinase II delta |
2511.89 nM 2511.89 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL3829 | Q13555 | CaM kinase II gamma |
3162.28 nM 3162.28 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL2828 | P48730 | Casein kinase I delta |
7943.28 nM 7943.28 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL5147 | P54760 | Ephrin type-B receptor 4 |
1000 nM |
IC50 |
PMID: 18849971
|
| CHEMBL203 | P00533 | Epidermal growth factor receptor erbB1 |
23 nM 1 nM 14.4 nM 3.1 nM 33 nM 54 nM 23 nM 80 nM 5.4 nM 39 nM 20 nM < 1 nM 33 nM 22 nM 33 nM 21 nM 33 nM 1 nM 33 nM 20 nM 20 nM 1 nM 70 nM 4.2 nM 1 nM 20 nM 7.3 nM 72 nM 33 nM 33 nM 0.1 nM 0.1 nM 62 nM 23 nM 39.9 nM 0.6 nM 1 nM 25 nM 1 nM 0.1 nM 20 nM 76 nM 0.45 nM 0.45 nM 26 nM < 1 nM 22 nM 22 nM 1.3 nM 15.5 nM 11.8 nM 0.585 nM 2.6 nM 0.509 nM 76 nM 70 nM 13 nM 0.1 nM |
IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 |
PMID: 11459659
PMID: 14684309 PMID: 16480284 PMID: 16480284 PMID: 16516473 PMID: 16516473 PMID: 16806916 PMID: 16806916 PMID: 17416531 PMID: 17889528 PMID: 17981366 PMID: 18849971 PMID: 19028424 PMID: 19170633 PMID: 19665377 PMID: 19969465 PMID: 20056425 PMID: 20166671 PMID: 20304535 PMID: 20550212 PMID: 20961149 PMID: 21334203 PMID: 21353546 PMID: 21724404 PMID: 22101132 PMID: 22119130 PMID: 22309911 PMID: 22818848 PMID: 23142066 PMID: 23611691 PMID: 23668441 PMID: 23792318 PMID: 23930994 PMID: 23973168 PMID: 23988354 PMID: 23988354 PMID: 24411123 PMID: 24411123 PMID: 24588073 PMID: 24607591 PMID: 24731281 PMID: 25409491 PMID: 25462282 PMID: 25468044 PMID: 26256032 PMID: 26275028 PMID: 26318056 PMID: 26451770 PMID: 26706113 PMID: 26829280 PMID: 26879314 PMID: 27132165 PMID: 27234887 PMID: 27288180 PMID: 27387355 DOI: 10.1039/C0MD00183J DOI: 10.1039/C5MD00208G via Super-PRED |
| CHEMBL1075167 | Q8NE63 | Homeodomain-interacting protein kinase 4 |
1258.93 nM 1258.93 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL3357 | P51617 | Interleukin-1 receptor-associated kinase 1 |
316.23 nM 316.23 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL3778 | Q9NWZ3 | Interleukin-1 receptor-associated kinase 4 |
1000 nM 1000 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL3836 | P53667 | LIM domain kinase 1 |
3981.07 nM 3981.07 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL260 | Q16539 | MAP kinase p38 alpha |
501.19 nM 501.19 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL4204 | Q9HBH9 | MAP kinase signal-integrating kinase 2 |
398.11 nM 398.11 nM 360 nM |
Ki Ki Kd |
via CMAUP
via CMAUP via Super-PRED |
| CHEMBL1293224 | P10636 | Microtubule-associated protein tau |
35481.3 nM 25118.9 nM 39810.7 nM |
Potency Potency Potency |
via CMAUP
via CMAUP via CMAUP |
| CHEMBL5518 | Q8N4C8 | Misshapen-like kinase 1 |
5011.87 nM 5011.87 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL6166 | O95819 | Mitogen-activated protein kinase kinase kinase kinase 4 |
1000 nM 1000 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL4852 | Q9Y4K4 | Mitogen-activated protein kinase kinase kinase kinase 5 |
1584.89 nM 1584.89 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL2349 | P15735 | Phosphorylase kinase gamma subunit 2 |
3162.28 nM 3162.28 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL1293235 | P02545 | Prelamin-A/C |
0.8 nM 0.8 nM |
Potency Potency |
via CMAUP
via Super-PRED |
| CHEMBL2595 | O94806 | Protein kinase C nu |
1258.93 nM 1258.93 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL1824 | P04626 | Receptor protein-tyrosine kinase erbB-2 |
240 nM 343 nM 599 nM 240 nM 240 nM 240 nM 280 nM 24 nM |
IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 |
PMID: 14684309
PMID: 16480284 PMID: 16480284 PMID: 20166671 PMID: 21334203 PMID: 22101132 PMID: 24411123 via Super-PRED |
| CHEMBL3009 | Q15303 | Receptor protein-tyrosine kinase erbB-4 |
158.49 nM 158.49 nM 476 nM |
Ki Ki IC50 |
via CMAUP
via CMAUP via Super-PRED |
| CHEMBL4202 | Q9H2G2 | Serine/threonine-protein kinase 2 |
398.11 nM 398.11 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL2527 | O96017 | Serine/threonine-protein kinase Chk2 |
630.96 nM 630.96 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL4900 | Q9BZL6 | Serine/threonine-protein kinase D2 |
2511.89 nM 2511.89 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL4355 | O14976 | Serine/threonine-protein kinase GAK |
7 nM |
Kd |
via Super-PRED
|
| CHEMBL2147 | P11309 | Serine/threonine-protein kinase PIM1 |
1995.26 nM 1995.26 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL5407 | Q86V86 | Serine/threonine-protein kinase PIM3 |
1258.93 nM 1258.93 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL5014 | O43353 | Serine/threonine-protein kinase RIPK2 |
530 nM |
Kd |
via Super-PRED
|
| CHEMBL5699 | Q9H0K1 | Serine/threonine-protein kinase SIK2 |
316.23 nM 316.23 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL1862 | P00519 | Tyrosine-protein kinase ABL |
1200 nM |
IC50 |
PMID: 18849971
|
| CHEMBL2250 | P51451 | Tyrosine-protein kinase BLK |
1000 nM 1000 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL4223 | P42685 | Tyrosine-protein kinase FRK |
251.19 nM 251.19 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL3234 | P08631 | Tyrosine-protein kinase HCK |
110 nM |
IC50 |
PMID: 18849971
|
| CHEMBL258 | P06239 | Tyrosine-protein kinase LCK |
398.11 nM 398.11 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL3905 | P07948 | Tyrosine-protein kinase Lyn |
158.49 nM 158.49 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL2041 | P07949 | Tyrosine-protein kinase receptor RET |
1700 nM |
IC50 |
PMID: 18849971
|
| CHEMBL4895 | P30530 | Tyrosine-protein kinase receptor UFO |
1584.89 nM 1584.89 nM |
Ki Ki |
via CMAUP
via CMAUP |
| CHEMBL267 | P12931 | Tyrosine-protein kinase SRC |
1100 nM |
IC50 |
PMID: 18849971
|
| CHEMBL279 | P35968 | Vascular endothelial growth factor receptor 2 |
14800 nM 14800 nM 48.5 nM |
IC50 IC50 IC50 |
PMID: 21353546
DOI: 10.1039/C0MD00183J via Super-PRED |
| CHEMBL1955 | P35916 | Vascular endothelial growth factor receptor 3 |
6309.57 nM 6309.57 nM |
Ki Ki |
via CMAUP
via CMAUP |
Predicted Targets (via Super-PRED):
| CHEMBL ID | UniProt ID | Name | Probability | Model accuracy |
|---|---|---|---|---|
| CHEMBL1293249 | Q13887 | Kruppel-like factor 5 | 97.91% | 86.33% |
| CHEMBL5747 | Q92793 | CREB-binding protein | 97.89% | 95.12% |
| CHEMBL225 | P28335 | Serotonin 2c (5-HT2c) receptor | 97.54% | 89.62% |
| CHEMBL2635 | P51452 | Dual specificity protein phosphatase 3 | 97.08% | 94.00% |
| CHEMBL2782 | P35610 | Acyl coenzyme A:cholesterol acyltransferase 1 | 95.39% | 91.65% |
| CHEMBL3251 | P19838 | Nuclear factor NF-kappa-B p105 subunit | 95.06% | 96.09% |
| CHEMBL2335 | P42785 | Lysosomal Pro-X carboxypeptidase | 95.05% | 100.00% |
| CHEMBL5608 | Q16288 | NT-3 growth factor receptor | 94.83% | 95.89% |
| CHEMBL4835 | P00338 | L-lactate dehydrogenase A chain | 94.06% | 95.34% |
| CHEMBL3976 | Q9UHL4 | Dipeptidyl peptidase II | 94.05% | 92.29% |
| CHEMBL5469 | Q14289 | Protein tyrosine kinase 2 beta | 93.51% | 91.03% |
| CHEMBL1937 | Q92769 | Histone deacetylase 2 | 93.17% | 94.75% |
| CHEMBL1980 | Q14524 | Sodium channel protein type V alpha subunit | 92.71% | 92.50% |
| CHEMBL4203 | Q9HAZ1 | Dual specificity protein kinase CLK4 | 92.65% | 94.45% |
| CHEMBL1075094 | Q16236 | Nuclear factor erythroid 2-related factor 2 | 92.35% | 96.00% |
| CHEMBL5925 | P22413 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 1 | 91.93% | 92.38% |
| CHEMBL2243 | O00519 | Anandamide amidohydrolase | 91.50% | 97.53% |
| CHEMBL240 | Q12809 | HERG | 91.20% | 89.76% |
| CHEMBL5339 | Q5NUL3 | G-protein coupled receptor 120 | 90.88% | 95.78% |
| CHEMBL3492 | P49721 | Proteasome Macropain subunit | 90.19% | 90.24% |
| CHEMBL202 | P00374 | Dihydrofolate reductase | 90.04% | 89.92% |
| CHEMBL3060 | Q9Y345 | Glycine transporter 2 | 89.91% | 99.17% |
| CHEMBL3401 | O75469 | Pregnane X receptor | 89.69% | 94.73% |
| CHEMBL2736 | Q14833 | Metabotropic glutamate receptor 4 | 89.45% | 97.56% |
| CHEMBL3108638 | O15164 | Transcription intermediary factor 1-alpha | 89.15% | 95.56% |
| CHEMBL2730 | P21980 | Protein-glutamine gamma-glutamyltransferase | 88.82% | 92.38% |
| CHEMBL344 | Q99705 | Melanin-concentrating hormone receptor 1 | 87.94% | 92.50% |
| CHEMBL5619 | P27695 | DNA-(apurinic or apyrimidinic site) lyase | 87.68% | 91.11% |
| CHEMBL235 | P37231 | Peroxisome proliferator-activated receptor gamma | 87.05% | 95.39% |
| CHEMBL4145 | Q9UKV0 | Histone deacetylase 9 | 86.95% | 85.49% |
| CHEMBL2373 | P21730 | C5a anaphylatoxin chemotactic receptor | 86.94% | 92.62% |
| CHEMBL1907601 | P11802 | Cyclin-dependent kinase 4/cyclin D1 | 86.17% | 98.99% |
| CHEMBL5163 | Q9NY46 | Sodium channel protein type III alpha subunit | 86.07% | 96.90% |
| CHEMBL1827 | O76074 | Phosphodiesterase 5A | 85.93% | 99.55% |
| CHEMBL3085 | P43003 | Excitatory amino acid transporter 1 | 85.06% | 94.67% |
| CHEMBL3524 | P56524 | Histone deacetylase 4 | 84.73% | 92.97% |
| CHEMBL5852 | Q96P65 | Pyroglutamylated RFamide peptide receptor | 83.33% | 85.00% |
| CHEMBL5845 | P23415 | Glycine receptor subunit alpha-1 | 83.28% | 90.71% |
| CHEMBL2047 | Q96RI1 | Bile acid receptor FXR | 83.02% | 96.10% |
| CHEMBL3038469 | P24941 | CDK2/Cyclin A | 83.00% | 91.38% |
| CHEMBL2535 | P11166 | Glucose transporter | 82.95% | 98.75% |
| CHEMBL290 | Q13370 | Phosphodiesterase 3B | 82.80% | 94.00% |
| CHEMBL1936 | P10721 | Stem cell growth factor receptor | 82.78% | 84.17% |
| CHEMBL2716 | Q8WUI4 | Histone deacetylase 7 | 82.69% | 89.44% |
| CHEMBL241 | Q14432 | Phosphodiesterase 3A | 82.20% | 92.94% |
| CHEMBL2007625 | O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | 82.10% | 99.00% |
| CHEMBL2424 | Q04760 | Glyoxalase I | 82.00% | 91.67% |
| CHEMBL1741221 | Q9Y4P1 | Cysteine protease ATG4B | 81.88% | 87.50% |
| CHEMBL3192 | Q9BY41 | Histone deacetylase 8 | 81.30% | 93.99% |
| CHEMBL2721 | P43005 | Excitatory amino acid transporter 3 | 81.06% | 93.50% |
| CHEMBL4208 | P20618 | Proteasome component C5 | 80.89% | 90.00% |
| CHEMBL4158 | P49327 | Fatty acid synthase | 80.81% | 82.50% |
Below are displayed all the plants proven (via scientific papers) to contain this
compound!
To see more specific details click the taxa you are interested in.
To see more specific details click the taxa you are interested in.
| Asarum heterotropoides |
| Asarum sieboldii |