| 10284-63-6 |
| Pinitol |
| 3-O-Methyl-D-chiro-inositol |
| Methylinositol |
| Inzitol |
| (+)-Pinitol |
| D-(+)-Pinitol |
| D-ononitol |
| Sennitol |
| (+)-Ononitol |
| 1-D-4-O-METHYL-MYO-INOSITOL |
| Matezit |
| Pinit |
| Ononitol |
| 484-68-4 |
| 6090-97-7 |
| D-chiro-Inositol, 3-O-methyl- |
| NIC5-15 |
| 1D-4-O-Methyl-myo-inositol |
| Ononitol, (+)- |
| 1D-3-O-methyl-chiro-inositol |
| Matezitol |
| Pinitol, (+)- |
| Piniol |
| Sennit |
| Cathartomannitol |
| Pinitol soy |
| Pinitol b |
| 4-O-Methyl-myo-inositol |
| DL-Pinitol |
| Inositol, 3-O-methyl-, D-chiro- |
| 5D-5-O-Methyl-chiro-inositol |
| D-Myo-inositol, 4-O-methyl- |
| (1S,2S,4S,5R)-6-methoxycyclohexane-1,2,3,4,5-pentol |
| UNII-TF9HZN9T0M |
| TF9HZN9T0M |
| UNII-N55OCE7X7M |
| N55OCE7X7M |
| UNII-A998ME07KR |
| D-3-o-methyl-chiro-inositol |
| A998ME07KR |
| CHEMBL493737 |
| CHEBI:28548 |
| (1R,2S,3S,4S,5S,6S)-6-Methoxycyclohexane-1,2,3,4,5-pentaol |
| NSC 43336 |
| NSC 128700 |
| NSC-128700 |
| chiro-Inositol, 3-O-methyl- |
| (1R,2S,3R,4S,5S,6S)-6-methoxycyclohexane-1,2,3,4,5-pentaol |
| (1R,2S,3R,4S,5S,6S)-6-methoxycyclohexane-1,2,3,4,5-pentol |
| (1R,2S,3S,4S,5S,6S)-6-methoxycyclohexane-1,2,3,4,5-pentol |
| (1r,2s,4s,5s)-6-methoxycyclohexane-1,2,3,4,5-pentol |
| Cathrtomannitol |
| NSC-43336 |
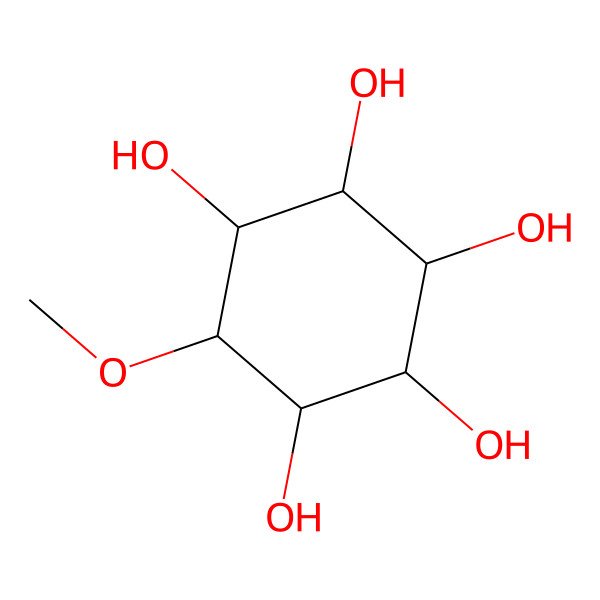
| O(C)C1C(O)C(O)C(O)C(O)C1O |
| 6-methoxycyclohexane-1,2,3,4,5-pentaol |
| MFCD00216659 |
| D-Pinitol, 95% |
| PINITOL [NDI] |
| PINITOL [MI] |
| PINITOL [USP-RS] |
| PINITOL [WHO-DD] |
| D0W0NU |
| METHYLINOSITOL [INCI] |
| 4-O-Methyl-D-chiro-inositol |
| (+/-)-PINITOL |
| NDI 18 [FDMS] |
| SCHEMBL464884 |
| D-Pinitol, analytical standard |
| METHYLINOSITOL, (+)- |
| NDI 18 |
| CHEMBL4303180 |
| SCHEMBL12858655 |
| SCHEMBL15269258 |
| CHEBI:18266 |
| DTXSID50883108 |
| DSCFFEYYQKSRSV-FEPQRWDDSA-N |
| DTXSID601029635 |
| DTXSID901337631 |
| METHYLINOSITOL RACEMATE[MI] |
| METHYLINOSITOL, (+/-)- |
| HY-N0655 |
| BDBM50275563 |
| s3870 |
| AKOS006287560 |
| AKOS015918369 |
| AKOS025310137 |
| BCP9000576 |
| CCG-266530 |
| DB12969 |
| 3-O-METHYL-(+)-CHIRO-INOSITOL |
| SMP2_000168 |
| D-chiro-Inositol, 3-O-methyl- (9CI) |
| AC-34710 |
| CS-0009678 |
| FT-0694792 |
| C03844 |
| C06352 |
| Q7094499 |
| W-203273 |
| B326207B-40FC-4D9A-B39B-15AE90545A34 |
| (1R,2S,3S,4S,5S,6S)-6-Methoxy-1,2,3,4,5-cyclohexanepentol |
| 3-O-METHYL-L,2,4 CIS-3,5,6 TRANS HEXAHYDROXYCYCLBHEXANOL |
|
There are more than 10 synonyms. If you wish to see them all click here.
|