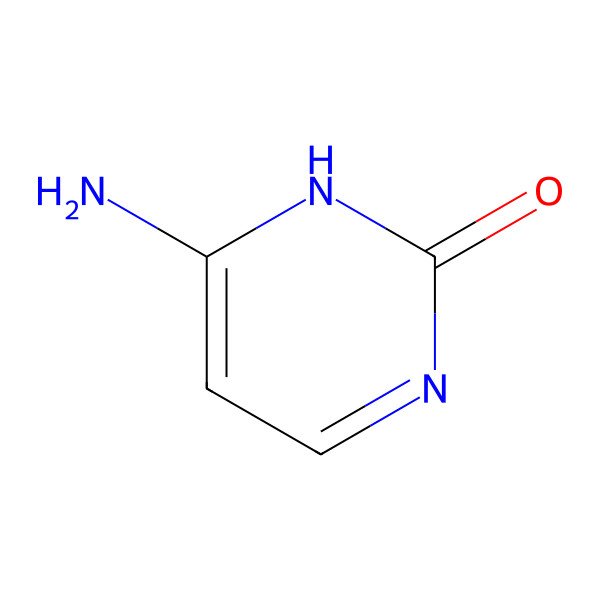
| 71-30-7 |
| 4-Amino-2-hydroxypyrimidine |
| Cytosinimine |
| 4-aminopyrimidin-2(1H)-one |
| 2(1H)-Pyrimidinone, 4-amino- |
| 4-Amino-2(1H)-pyrimidinone |
| 6-Aminopyrimidin-2(1h)-One |
| Cytosin |
| 4-aminopyrimidin-2-ol |
| Zytosin |
| 6-amino-1H-pyrimidin-2-one |
| 4-amino-2-oxo-1,2-dihydropyrimidine |
| Cyt |
| AI3-52281 |
| MFCD00006034 |
| 4-Amino-1H-pyrimidin-2-one |
| CHEBI:16040 |
| 2(1H)-pyrimidinone, 6-amino- |
| EINECS 200-749-5 |
| UNII-8J337D1HZY |
| NSC 27787 |
| NSC-27787 |
| 8J337D1HZY |
| 107646-83-3 |
| DTXSID4044456 |
| 134434-40-5 |
| 107646-84-4 |
| 134434-39-2 |
| 2(1H)-Pyrimidinone, 3,4-dihydro-4-imino-, (E)- (9CI) |
| 4-Aminopyrimidin-2-(1H)-one |
| DTXCID2024456 |
| EC 200-749-5 |
| NSC27787 |
| 2(1H)-Pyrimidinone, 3,4-dihydro-4-imino-, (Z)- (9CI) |
| 4-Aminopyrimidin-2(1H)-one (Cytosine) |
| CYTOSINE (USP-RS) |
| CYTOSINE [USP-RS] |
| SMR000857094 |
| LAMIVUDINE IMPURITY E (EP IMPURITY) |
| LAMIVUDINE IMPURITY E [EP IMPURITY] |
| LAMIVUDINE IMPURITY C (USP IMPURITY) |
| LAMIVUDINE IMPURITY C [USP IMPURITY] |
| 4-amino-2-pyrimidinol |
| aminopyrimidone |
| iminopyrimidinone |
| 3h-cytosine |
| Cytosine (8CI) |
| 4-amino-1,2-dihydropyrimidin-2-one |
| Lamivudine impurity c |
| 2-Pyrimidinol, 1,4-dihydro-4-imino-, (Z)- (9CI) |
| Gemcitabine impurity A |
| Cytosine, >=99% |
| 2(1H)-Pyrimidinone, 4-amino- (9CI) |
| CYTOSINE [INCI] |
| Lamivudine impurity c rs |
| CYTOSINE [MI] |
| 4-amino-pyrimidin-2-ol |
| 4-Amino-2-oxypyrimidine |
| bmse000180 |
| CYTOSINE [WHO-DD] |
| D0G7IR |
| Epitope ID:167475 |
| 4-Amino-2(1H)pyrimidone |
| SCHEMBL4059 |
| 4-Amino-2(1H)-pyrimidone |
| 2-Hydroxy-6-amino-pyrimidin |
| 4-amino-3h-pyrimidin-2-one |
| MLS001332635 |
| MLS001332636 |
| CHEMBL15913 |
| 6-Amino-2(1H)-pyrimidinone |
| 2-Pyrimidinol, 1,6-dihydro-6-imino-, (E)- (9CI) |
| GTPL8490 |
| HMS2233N21 |
| HMS3369N05 |
| Cytosine, >=99.0% (HPLC) |
| BCP22793 |
| HY-I0626 |
| STR01426 |
| Tox21_302139 |
| s4893 |
| STK366767 |
| STL455080 |
| 6-amino-1,2-dihydropyrimidin-2-one |
| AKOS000120336 |
| AKOS005443393 |
| AKOS015896942 |
| AC-2489 |
| AM83918 |
| BCP9000005 |
| CCG-266052 |
| CS-W020703 |
| 6-amino-1H-pyrimidin-2-one;CYTOSINE |
| CAS-71-30-7 |
| CID 5274263 |
| SRI-2354-05 |
| NCGC00247019-01 |
| NCGC00255926-01 |
| BP-20183 |
| Cytosine, Vetec(TM) reagent grade, 99% |
| NCI60_012445 |
| SY001643 |
| 4-imino-3,4-dihydropyrimidin-2(1H)-one |
| LS-135824 |
| FT-0617471 |
| EN300-21504 |
| C00380 |
| 2-Pyrimidinol,1,6-dihydro-6-imino-,(E)-(9ci) |
| A837149 |
| Q178425 |
| 2(1H)-Pyrimidinone,3,4-dihydro-4-imino-,(E)-(9ci) |
| CBA1D098-C5AB-46CE-AAC6-754572886EB2 |
| Z203045338 |
| Cytosine, United States Pharmacopeia (USP) Reference Standard |
| Cytosine, Pharmaceutical Secondary Standard; Certified Reference Material |
| Gemcitabine impurity A, European Pharmacopoeia (EP) Reference Standard |
|
There are more than 10 synonyms. If you wish to see them all click here.
|