| Phomin |
| 14930-96-2 |
| cytochalasin-B |
| CHEBI:23527 |
| C29H37NO5 |
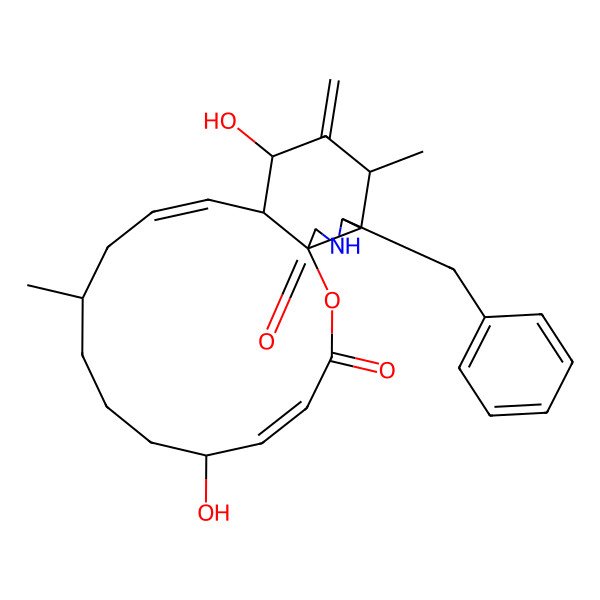
| Cytochalasin B (Phomin) |
| UNII-3CHI920QS7 |
| MLS000028816 |
| 3CHI920QS7 |
| CCRIS 9284 |
| NSC-107658 |
| SMR000058787 |
| Cytochalasin beta,helminthosporium dematioideum |
| (1S,4E,6R,10R,12E,14S,15S,17S,18S,19S)-19-benzyl-6,15-dihydroxy-10,17-dimethyl-16-methylidene-2-oxa-20-azatricyclo[12.7.0.01,18]henicosa-4,12-diene-3,21-dione |
| HSDB 3479 |
| EINECS 239-000-2 |
| 24-Oxa(14)cytochalasa-6(12),13,21-triene-1,23-dione, 7,20-dihydroxy-16-methyl-10-phenyl-, (7S,13E,16R,20R,21E)- |
| BRN 1096207 |
| (4Z,12Z)-19-Benzyl-6,15-dihydroxy-10,17-dimethyl-16-methylidene-2-oxa-20-azatricyclo[12.7.0.01,18]henicosa-4,12-diene-3,21-dione |
| 2H-Oxacyclotetradecino(2,3-d)isoindole-2,18(5H)-dione, 6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-5,13-dihydroxy-9,15-dimethyl-14-methylene-16-(phenylmethyl)-, (3E,5R,9R,11E,12aS,13S,15S,15aS,16S,18aS)- |
| MLS002703001 |
| Biostent |
| Preverex |
| C29-H37-N-O5 |
| NSC107658 |
| 7,20-Dihydroxy-10-phenyl-5,16-dimethyl-24-oxa-(14)cytochalas-6(12),13,21-trien-23-one |
| (E,E)-(5S,9R,12aS,13S,15S,15aS,16S,18aS)-16-Benzyl-6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-5,13-dihydroxy-9,15-dimethyl-14-methylene-2H-oxacyclotetradec(2,3-d)isoindole-2,18(5H)-dione |
| 2H-Oxacyclotetradec(2,3-d)isoindole-2,18(5H)-dione, 16-benzyl-6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-5,13-dihydroxy-9,15-dimethyl-14-methylene-, (E,E)-(5S,9R,12aS,13S,15S,15aS,16S,18aS)- |
| MFCD00077704 |
| ROTHWEILER |
| Cytochalasin B from Drechslera dematioidea |
| Opera_ID_1877 |
| Spectrum5_001780 |
| D0DJ4A |
| CYTOCHALASIN B [MI] |
| BSPBio_001600 |
| MLS001148651 |
| CYTOCHALASIN B [HSDB] |
| Cytochalasin B containing stent |
| CHEMBL411729 |
| GTPL5334 |
| SCHEMBL20063279 |
| BCBcMAP01_000011 |
| DTXSID30893482 |
| GBOGMAARMMDZGR-TYHYBEHESA-N |
| REGID_for_CID_5311281 |
| HMS1361P22 |
| HMS1791P22 |
| HMS1989P22 |
| HMS3402P22 |
| 24-Oxa[14]cytochalasa-6(12),13,21-triene-1,23-dione, 7,20-dihydroxy-16-methyl-10-phenyl-, (7S,13E,16R,20R,21E)- |
| 2H-Oxacyclotetradec(2,3-d)isoindole-2,18(5H)-dione, 16-benzyl-6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-5,13-dihydroxy-9,15-dimethyl-14-methylene-, (E)-(5S,9R,12aS,13S,15S,15aS,16aS,18aS)- |
| 2H-Oxacyclotetradecino(2,3-d)isoindole-2,18(5H)-dione, 16-benzyl-6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-5,13-dihydroxy-9,15-dimethyl-14-methylene-, (E,E)-(5S,9R,12aS,13S,15S,15aS,16S,18aS)- |
| 7(S),20(R)-Dihydroxy-16(R)-methyl-10-phenyl-24-oxa(14)cytochalasa-6(12),13(E),21(E)-triene-1,23-dione |
| 7(S),20(R)-dihydroxy-16(R)-methyl-10-phenyl-24-oxa[14]cytochalasa-6(12),13(E),21(E)-triene-1,23-dione |
| BDBM50478403 |
| CCG-39777 |
| LMPK11000002 |
| AKOS025311470 |
| IDI1_034070 |
| NCGC00163439-01 |
| NCGC00163439-02 |
| HY-16928 |
| CS-0012975 |
| C19954 |
| Cytochalasin B from Helminthosporium dematioideum |
| W-201323 |
| Q26998028 |
| Cytochalasin B from Drechslera dematioidea, >=97.0% (HPLC) |
| Cytochalasin B from Drechslera dematioidea, >=98% (HPLC), powder |
| (3E,5R,9R,11E,12aS,13S,15S,15aS,16S,18aS)-16-benzyl-5,13-dihydroxy-9,15-dimethyl-14-methylidene-6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-2H-oxacyclotetradecino[2,3-d]isoindole-2,18(5H)-dione |
| (3E,5R,9R,11E,12aS,13S,15S,15aS,16S,18aS)-6,7,8,9,10,12a,13,14,15,15a,16,17-Dodecahydro-5,13-dihydroxy-9,15-dimethyl-14-methylene-16-(phenylmethyl)-2H-oxacyclotetradecino[2,3-d]isoindole-2,18(5H)-dione |
| (3S,3aS,4S,6S,6aS,7E,10R,14R,15E,181S)-3-benzyl-6,14-dihydroxy-4,10-dimethyl-5-methylene-3,3a,4,5,6,6a,9,10,11,12,13,14-dodecahydro-1H-[1]oxacyclotetradeca[2,3-d]isoindole-1,17(2H)-dione |
| 11032-95-4 |
| 2H-OXACYCLOTETRADECINO(2,3-D)ISOINDOLE-2,18(5H)-DIONE, 16-BENZYL-6,7,8,9,10,12A,13,14,15,15A,16,17-DODECAHYDRO-5,13-DIHYDROXY-9,15-DIMETHYL-14-METHYLENE-, (E,E)-(5R,9R,12AS,13S,15S,15AS,16S,18AS)- |
| 2H-OXACYCLOTETRADECINO(2,3-D)ISOINDOLE-2,18(5H)-DIONE, 6,7,8,9,10,12A,13,14,15,15A,16,17-DODECAHYDRO-5,13-DIHYDROXY-9,15-DIMETHYL-14-METHYLENE-16-(PHENYLMETHYL)-, (5R-(3E,5R*,9R*,11E,12AS*,13S*,15S*,15AS*,16S*,18AS*))- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|