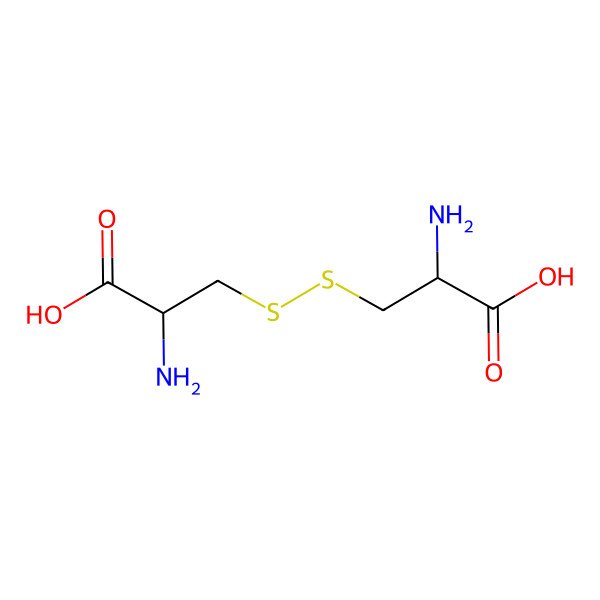
| 56-89-3 |
| cystine |
| L-Cystin |
| L-Dicysteine |
| Cysteine disulfide |
| Cystin |
| beta,beta'-Dithiodialanine |
| L-Cysteine disulfide |
| 1-Cystine |
| 3,3'-Dithiodialanine |
| Cystine, L- |
| Dicysteine |
| Cystine (L)- |
| Cystine acid |
| (H-Cys-OH)2 |
| Alanine, 3,3'-dithiodi- |
| Alanine, 3,3'-dithiobis- |
| beta,beta'-Dithioalanine, L- |
| L-Alanine, 3,3'-dithiobis- |
| 3,3'-Dithiobis(2-aminopropanoic acid) |
| Cystine (VAN) |
| Cystine [USAN] |
| Cystin (VAN) |
| L-(-)-Cystine |
| (-)-Cystine |
| Gelucystine |
| CCRIS 5822 |
| L-alpha-Diamino-beta-dithiolactic acid |
| Cystine,d |
| Bis(beta-amino-beta-carboxyethyl)disulfide |
| UNII-48TCX9A1VT |
| 3,3'-Dithiobis-L-alanine |
| EINECS 200-296-3 |
| (R-(R*,R*))-3,3'-Dithiobis(2-aminopropanoic acid) |
| 48TCX9A1VT |
| NSC 13203 |
| beta,beta'-Diamino-beta,beta'-dicarboxydiethyl disulfide |
| BRN 1728094 |
| DTXSID2046418 |
| CHEBI:16283 |
| AI3-09064 |
| beta,beta'-Diamino-beta,beta'-dicarboxydiethyldisulfide |
| Cystine [USAN:INN] |
| (2R,2'R)-3,3'-disulfanediylbis(2-aminopropanoic acid) |
| 3,3'-Dithiobis(2-aminopropanoic acid), (R-(R*,R*))- |
| Propanoic acid, 3,3'-dithiobis(2-amino-, (R-(R*,R*))- |
| NSC-13203 |
| S-CYSTEINYL CYSTEINE |
| Bis(beta-amino-beta-carboxyethyl) disulfide |
| DTXCID0026418 |
| EC 200-296-3 |
| (2R)-2-amino-3-{[(2R)-2-amino-2-carboxyethyl]disulfanyl}propanoic acid |
| 4-04-00-03155 (Beilstein Handbook Reference) |
| Cystine (L-Cystine) |
| C6H12N2O4S2 |
| E921 |
| CYSTINE (MART.) |
| CYSTINE [MART.] |
| CYSTINE (USP-RS) |
| CYSTINE [USP-RS] |
| (R,R)-3,3'-dithiobis(2-aminopropanoic acid) |
| S-(((R)-2-amino-2-carboxyethyl)thio)cysteine |
| cistina |
| Zystin |
| cystin- |
| CYSTINE (EP MONOGRAPH) |
| CYSTINE [EP MONOGRAPH] |
| 3,3-DISULFANEDIYLBIS((2R)-2-AMINOPROPANOIC ACID) |
| 2-amino-3-(2-amino-2-carboxy-ethyl)disulfanyl-propanoic acid |
| L-Cystin- |
| alpha-Diamino-beta-dithiolactic acid |
| (R-(R*,R*))-3,3'-Dithiobis |
| [R-(R*,R*)]-3,3'-Dithiobis |
| MFCD00064228 |
| L Cystine |
| (2R)-2-amino-3-[[(2R)-2-amino-2-carboxyethyl]disulfanyl]propanoic acid |
| CAS-56-89-3 |
| 2079930-29-1 |
| TYROSINE IMPURITY C (EP IMPURITY) |
| TYROSINE IMPURITY C [EP IMPURITY] |
| (R,R)-3,3'-Dithiobis(2-aminopropionicacid) |
| ACETYLCYSTEINE IMPURITY A (EP IMPURITY) |
| ACETYLCYSTEINE IMPURITY A [EP IMPURITY] |
| 24645-67-8 |
| [R-(R*,R*)]-3,3'-dithiobis[2-aminopropanoic acid] |
| cystinum |
| DTXSID50859005 |
| C6-H12-N2-O4-S2 |
| NCGC00164531-01 |
| IYY |
| 3,3'-Dithiobis |
| b,b'-Dithiodialanine |
| L-Cystine (9CI) |
| Cystine (USAN/INN) |
| L-Cystine (JP17) |
| CYSTINE [VANDF] |
| CYSTINE [INCI] |
| CYSTINE [INN] |
| CYSTINE [MI] |
| L-CYSTINE [FCC] |
| L-CYSTINE [JAN] |
| Cystine, L- (8CI) |
| CYSTINE [WHO-DD] |
| bmse000035 |
| D0V1CP |
| (2R)-2-amino-3-[[(2R)-2-amino-2-carboxy-ethyl]disulfanyl]propanoic acid |
| beta,beta'-Dithiobisalanine |
| SCHEMBL10226 |
| L-Cystine, non-animal source |
| CHEMBL590540 |
| GTPL5413 |
| (H-Cys-OH)2 (Disulfide bond) |
| DTXCID60210087 |
| HY-N0394 |
| Tox21_112162 |
| CCG-36355 |
| L-Cystine, >=99.7% (TLC) |
| s4808 |
| Bis(b-amino-b-carboxyethyl) disulfide |
| Cystine, NIST(R) SRM(R) 143d |
| AKOS015898645 |
| Tox21_112162_1 |
| AM81644 |
| DB00138 |
| (2R)-2-azanyl-3-[[(2R)-2-azanyl-3-oxidanyl-3-oxidanylidene-propyl]disulfanyl]propanoic acid |
| Bis(b-amino-beta-carboxyethyl) disulfide |
| NCGC00166006-01 |
| NCGC00166006-02 |
| AC-11189 |
| AS-12654 |
| D(+)-3,3'-Dithiobis(2-aminopropanoate |
| L-Cystine, >=98% (TLC), crystalline |
| L-Cystine, BioUltra, >=99.5% (T) |
| LS-59051 |
| L-Cystine, SAJ special grade, >=99.0% |
| L-Cystine, Vetec(TM) reagent grade, 98% |
| ACETYLCYSTEINE IMPURITY A (L-CYSTINE) |
| b,b'-Diamino-b,b'-dicarboxydiethyl disulfide |
| CS-0008930 |
| D(+)-3,3'-Dithiobis(2-aminopropanoic acid |
| (R,R)-3,3'-Dithiobis(2-aminopropionic Acid) |
| C00491 |
| D03636 |
| EN300-174654 |
| M06021 |
| Q408626 |
| 3,3'-Disulfanediylbis((2R)-2-aminopropanoic acid) |
| 3,3'-dithiobis[2-amino-[R-(R*,R*)]-Propanoate |
| 2-Amino-3-[(2-amino-2-carboxyethyl)dithio]propanoate |
| L-Cystine, certified reference material, TraceCERT(R) |
| 3,3'-dithiobis[2-amino-[R-(R*,R*)]-Propanoic acid |
| Cystine, European Pharmacopoeia (EP) Reference Standard |
| Z1269145231 |
| 2-amino-3-(2-amino-2-carboxy-ethyl)disulfanyl-propanoate |
| 2-Amino-3-[(2-amino-2-carboxyethyl)dithio]propanoic acid |
| DD82F461-3F8F-4624-9E2C-0272A9FA79ED |
| Cystine, United States Pharmacopeia (USP) Reference Standard |
| L-Cystine, Cell Culture Reagent (H-L-Cys(1)-OH.H-L-Cys(1)-OH) |
| L-Cystine, Pharmaceutical Secondary Standard; Certified Reference Material |
| L-Cystine, produced by Wacker Chemie AG, Burghausen, Germany, >=98.5% |
| L-Cystine, from non-animal source, meets EP testing specifications, suitable for cell culture, 98.5-101.0% |
|
There are more than 10 synonyms. If you wish to see them all click here.
|