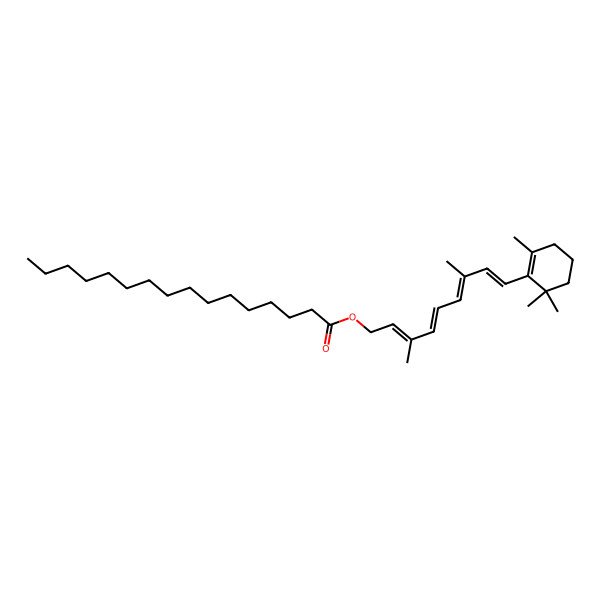
| VITAMIN A PALMITATE |
| 79-81-2 |
| Retinol palmitate |
| Retinol, hexadecanoate |
| all-trans-Retinyl palmitate |
| Arovit |
| Optovit-A |
| Retinyl hexadecanoate |
| Aquapalm |
| Dispatabs Tabs |
| Vitazyme A |
| Optovit A |
| Axerophthol palmitate |
| Vitamin A Solubilized |
| trans-Retinol palmitate |
| trans-Retinyl palmitate |
| Retinyl (Vitamin A) Palmitate |
| Lutavit A 500 Plus |
| all-trans-Retinol palmitate |
| [(2E,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethylcyclohexen-1-yl)nona-2,4,6,8-tetraenyl] hexadecanoate |
| C36H60O2 |
| Arovit (Roche) |
| all-trans-Vitamin A palmitate |
| Aquasol A (TN) |
| CCRIS 3280 |
| Retinol, palmitate, all-trans- |
| Retinol palmitate [JAN] |
| EINECS 201-228-5 |
| O~15~-hexadecanoylretinol |
| UNII-1D1K0N0VVC |
| NSC-758478 |
| BRN 1917366 |
| 1D1K0N0VVC |
| Retinol, palmitate, all-trans |
| Retinol palmitate (6CI,7CI) |
| Retinol, all-trans-, palmitate |
| DTXSID1021241 |
| CHEBI:17616 |
| Palmitic acid, ester with retinol |
| all-trans-RetinylPalmitate-d5(Major) |
| Retinol, palmitate, all-trans- (8CI) |
| NCGC00095056-03 |
| EC 201-228-5 |
| (2E,4E,6E,8E)-Hexadecanoic acid 3,7-dimethyl-9-(2,6,6-trimethyl-cyclohex-1-enyl)-nona-2, 4,6,8,tetraenyl ester |
| DTXCID101241 |
| Retinyl (palmitate) |
| CAS-79-81-2 |
| (2E,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethylcyclohex-1-en-1-yl)nona-2,4,6,8-tetraen-1-yl hexadecanoate |
| SMR000112463 |
| retinylpalmitate |
| retinol-palmitate |
| retinyl-palmitate |
| Vitamin A palmitate;Retinol palmitate |
| 1331666-45-5 |
| Palmitic acid retinol |
| Retinyl palmitic acid |
| Vitamin- A palmitate |
| Retinol, hexadecanoato |
| O~15~-palmitoylretinol |
| Retinyl hexadecanoic acid |
| Spectrum5_001201 |
| All-(e)-retinol palmitate |
| bmse000501 |
| Retinol palmitate (JP17) |
| CHEMBL1675 |
| SCHEMBL41649 |
| MLS001332437 |
| MLS001332438 |
| SPECTRUM1503604 |
| all-trans-retinyl hexadecanoate |
| RETINYL PALMITATE [INCI] |
| HMS500M11 |
| RETINYL PALMITATE [VANDF] |
| VITAMIN A PALMITATE [MI] |
| Vitamin a palmitate (solubilized) |
| HMS1922E10 |
| HMS2093G13 |
| HMS2268C06 |
| Pharmakon1600-01503604 |
| RETINOL PALMITATE [WHO-DD] |
| RETINYL PALMITATE [USP-RS] |
| AMY13840 |
| HY-B1384 |
| VAE 16:0 |
| VITAMIN A PALMITATE [VANDF] |
| Tox21_113452 |
| Tox21_303008 |
| CCG-39342 |
| LMPR01090013 |
| MFCD00019414 |
| NSC758478 |
| s4126 |
| retinol, O~15~-(1-oxohexadecyl)- |
| AKOS015918435 |
| C36-H60-O2 |
| CS-O-02552 |
| LS-2307 |
| NSC 758478 |
| Vitamin A palmitate, 1.7 M.I.U./g |
| IDI1_000249 |
| NCGC00095056-01 |
| NCGC00095056-02 |
| NCGC00256427-01 |
| AC-20001 |
| MS-29721 |
| VITAMIN A PALMITATE [ORANGE BOOK] |
| SBI-0051830.P002 |
| ALL-(E)-RETINOL PALMITATE [WHO-IP] |
| CS-0013116 |
| C02588 |
| C76552 |
| D00164 |
| AB00052360_04 |
| A839762 |
| SR-05000001910 |
| VITAMIN A PALMITATE (SOLUBILIZED) [VANDF] |
| Q7316807 |
| SR-05000001910-1 |
| Retinyl palmitate (Vitamin A palmitate; Retinol palmitate) |
| Retinyl palmitate, potency: >=1,700,000 USP units per g |
| VITAMIN A (AS PALMITATE & BETA CAROTENE) [VANDF] |
| Retinyl palmitate, United States Pharmacopeia (USP) Reference Standard |
| [3,7-dimethyl-9-(2,6,6-trimethylcyclohexen-1-yl)nona-2,4,6,8-tetraenyl] hexadecanoate |
| 3,7-Dimethyl-9-(2,6,6,-trimethyl-1-cyclohexen-1-yl)-2,4,6,8-nonatetraen-1-ol palmitate |
| (2E,4E,6E,8E)-3,7-Dimethyl-9-(2,6,6-trimethyl-cyclohex-en-1-yl)-2,4,6,8-nonateetraen-1-yl-palmitate |
| (2E,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethylcyclohex-1-enyl)nona-2,4,6,8-tetraenyl palmitate |
| 110067-62-4 |
| 37340-08-2 |
| hexadecanoic acid [(2E,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethyl-1-cyclohexenyl)nona-2,4,6,8-tetraenyl] ester |
| Retinyl Palmitate (Vitamin A Palmitate), Pharmaceutical Secondary Standard; Certified Reference Material |
|
There are more than 10 synonyms. If you wish to see them all click here.
|