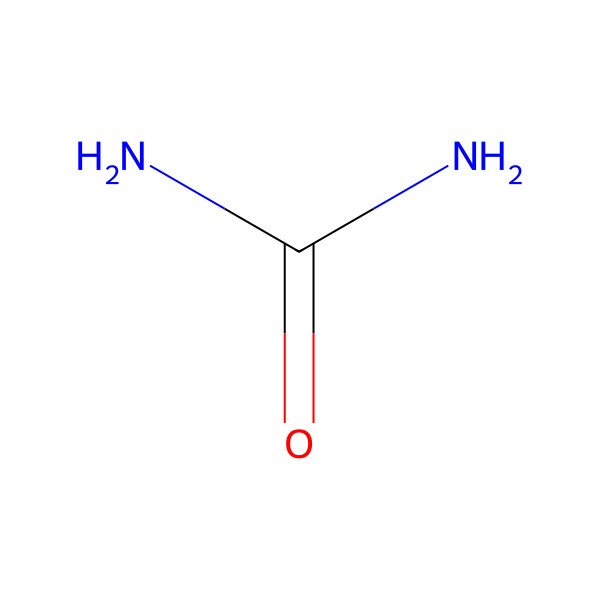
| carbamide |
| 57-13-6 |
| Carbonyldiamide |
| Isourea |
| Ureophil |
| Carbonyldiamine |
| Ureaphil |
| Carbamimidic acid |
| Urevert |
| Alphadrate |
| Aquadrate |
| Carbaderm |
| Keratinamin |
| Pseudourea |
| Carbonyl diamide |
| Calmurid |
| Pastaron |
| Urepearl |
| Carbamide resin |
| Ultra Mide |
| Varioform ii |
| Aqua Care |
| carmol |
| Harnstoff |
| Mocovina |
| Nutraplus |
| Prespersion, 75 urea |
| B-I-K |
| Supercel 3000 |
| Aqua Care HP |
| Ureacin-10 lotion |
| Ureacin-20 |
| Ureacin-40 Creme |
| Caswell No. 902 |
| Hyanit |
| Carbamimic acid |
| NCI-C02119 |
| Antisepsis bolus |
| Elaqua xx |
| Sterile Urea |
| Carbonyl Diamine |
| Keratinamin Kowa |
| CCRIS 989 |
| HSDB 163 |
| Mocovina [Czech] |
| Benural 70 |
| Harnstoff [German] |
| aminoketone |
| NSC 34375 |
| Urea [JAN] |
| uree |
| Isoharnstoff |
| Aquacare |
| Karbamid |
| Urepeal |
| EPA Pesticide Chemical Code 085702 |
| Urea perhydrate |
| Bubber shet |
| Urepeal L |
| AI3-01202 |
| Azodicarboxylic acid-diamide |
| EINECS 200-315-5 |
| UR |
| NSC-34375 |
| Pastaron 10 |
| Pastaron 20 |
| Pastaron 20 soft |
| INS NO.927A |
| carbonamidimidic acid |
| E927b |
| INS-927A |
| 37955-36-5 |
| DTXSID4021426 |
| UNII-8W8T17847W |
| CHEBI:16199 |
| Urea-18O |
| Urea [USP:JAN] |
| Carbamide;Carbonyldiamide |
| E-927A |
| MFCD00008022 |
| 8W8T17847W |
| H2NC(O)NH2 |
| H2N-C(OH)=NH |
| U-CORT COMPONENT UREA |
| H2N-C(=NH)-OH |
| HO-C(=NH)-NH2 |
| (NH2)2CO |
| ALPHADERM COMPONENT UREA |
| DTXCID901426 |
| 4744-36-9 |
| CARMOL HC COMPONENT UREA |
| Helicosol |
| UREA COMPONENT OF U-CORT |
| CHEBI:48376 |
| CALMURID HC COMPONENT UREA |
| EC 200-315-5 |
| UREA COMPONENT OF ALPHADERM |
| UREA COMPONENT OF CARMOL HC |
| NSC34375 |
| UREA COMPONENT OF CALMURID HC |
| NCGC00090892-01 |
| UREA (II) |
| UREA (MART.) |
| UREA [II] |
| UREA [MART.] |
| UREA (EP MONOGRAPH) |
| UREA (USP IMPURITY) |
| UREA [EP MONOGRAPH] |
| UREA [USP IMPURITY] |
| UREA (USP MONOGRAPH) |
| UREA [USP MONOGRAPH] |
| 3138-51-0 |
| URE |
| Ureum |
| CAS-57-13-6 |
| ALLANTOIN IMPURITY B (EP IMPURITY) |
| ALLANTOIN IMPURITY B [EP IMPURITY] |
| FLUOROURACIL IMPURITY G (EP IMPURITY) |
| FLUOROURACIL IMPURITY G [EP IMPURITY] |
| UREA, ACS |
| isoureas |
| Lanaphilic |
| Cerovel |
| Uroderm |
| Vanamide |
| amino ketone |
| amino-ketone |
| Pastaron soft |
| Urea, ultrapure |
| E-Cardamoni |
| Aquacare HP |
| Carbamide solution |
| beta-I-k |
| Pastaron (TN) |
| Carmol 40 |
| NIMIN |
| Cerovel (Salt/Mix) |
| Panafil (Salt/Mix) |
| Rubinol ST 010 |
| Urea, p.a. |
| Optigen 1200 |
| Urea, 2M |
| Urea,(S) |
| Spectrum_000672 |
| URE (CHRIS Code) |
| UREA [VANDF] |
| WLN: ZVZ |
| Carbamimidic acid (VAN) |
| Spectrum2_001192 |
| Spectrum3_001791 |
| Spectrum4_001168 |
| Spectrum5_001862 |
| Urea (JP17/USP) |
| Urea, analytical standard |
| Urea, for electrophoresis |
| UREA [HPUS] |
| UREA [HSDB] |
| UREA [INCI] |
| Urea (8CI,9CI) |
| UREA [FCC] |
| UREA [USP-RS] |
| UREA [WHO-DD] |
| Eucerin 10% Urea Lotion |
| CHEMBL985 |
| D02XBW |
| UREA [MI] |
| UREA [ORANGE BOOK] |
| H2N-CO-NH2 |
| Urea A.C.S. reagent grade |
| Pesticide Code: 085702 |
| BSPBio_003341 |
| KBioGR_001775 |
| KBioSS_001152 |
| Urea, puriss., 99.5% |
| MLS001076688 |
| DivK1c_000086 |
| SPECTRUM1500604 |
| Urea, AR, >=99% |
| Urea, LR, >=99% |
| SPBio_001263 |
| GTPL4539 |
| Urea, >=99.0% |
| Urea, Molecular Biology Reagent |
| BDBM24961 |
| CHEBI:48379 |
| HMS500E08 |
| KBio1_000086 |
| KBio2_001152 |
| KBio2_003720 |
| KBio2_006288 |
| KBio3_002843 |
| (C-H4-N2-O)x- |
| Urea 100 microg/mL in Methanol |
| NINDS_000086 |
| HMS1921I17 |
| HMS2092C08 |
| HMS2232P21 |
| Pharmakon1600-01500604 |
| component of Artra Ashy Skin Cream |
| AMY37159 |
| BCP30439 |
| CS-B1800 |
| HY-Y0271 |
| STR00449 |
| Urea, tested according to Ph.Eur. |
| Tox21_111036 |
| Tox21_202158 |
| Tox21_300035 |
| c0165 |
| CCG-40265 |
| NSC757375 |
| s3687 |
| STL194286 |
| Urea, ReagentPlus(R), >=99.5% |
| AKOS009031424 |
| Urea, NIST(R) SRM(R) 2141 |
| Urea, SAJ first grade, >=98.0% |
| DB03904 |
| LS-1389 |
| NSC-757375 |
| Urea, JIS special grade, >=99.0% |
| Urea, meets USP testing specifications |
| Urea, Vetec(TM) reagent grade, 99% |
| IDI1_000086 |
| Basodexan pound>>Carmol pound>>Carbamide |
| NCGC00090892-02 |
| NCGC00090892-03 |
| NCGC00090892-04 |
| NCGC00090892-05 |
| NCGC00090892-06 |
| NCGC00254181-01 |
| NCGC00259707-01 |
| SMR000499585 |
| Urea, ACS reagent, 99.0-100.5% |
| 4,4-(3-Oxapentanediyldioxy)dibenzaldehyde |
| SBI-0051552.P002 |
| Urea, BioReagent, suitable for cell culture |
| FT-0645129 |
| FT-0675737 |
| FT-0675738 |
| U0073 |
| U0077 |
| Urea, ReagentPlus(R), >=99.5%, pellets |
| EN300-19456 |
| C00086 |
| component of Artra Ashy Skin Cream (Salt/Mix) |
| D00023 |
| D70446 |
| M02656 |
| Q48318 |
| Urea, p.a., ACS reagent, 99.0-100.5% |
| AB00052123_05 |
| SR-01000762961 |
| Urea, NIST(R) SRM(R) 912a, clinical standard |
| InChI=1/CH4N2O/c2-1(3)4/h(H4,2,3,4 |
| SR-01000762961-2 |
| Urea, British Pharmacopoeia (BP) Reference Standard |
| Urea, European Pharmacopoeia (EP) Reference Standard |
| F0001-1490 |
| Urea, BioUltra, for molecular biology, >=99.5% (T) |
| Urea, BioXtra, pH 7.5-9.5 (20 C, 5 M in H2O) |
| Urea, NIST SRM 2152, combustion calorimetric standard |
| Urea, United States Pharmacopeia (USP) Reference Standard |
| 6E4EB293-4363-4D38-BF3B-1397372C31E5 |
| Urea, 8 M (after reconstitution with 16 mL high purity water) |
| Urea, puriss. p.a., ACS reagent, reag. Ph. Eur., >=99.5% |
| Urea, Pharmaceutical Secondary Standard; Certified Reference Material |
| Urea, powder, BioReagent, for molecular biology, suitable for cell culture |
|
There are more than 10 synonyms. If you wish to see them all click here.
|