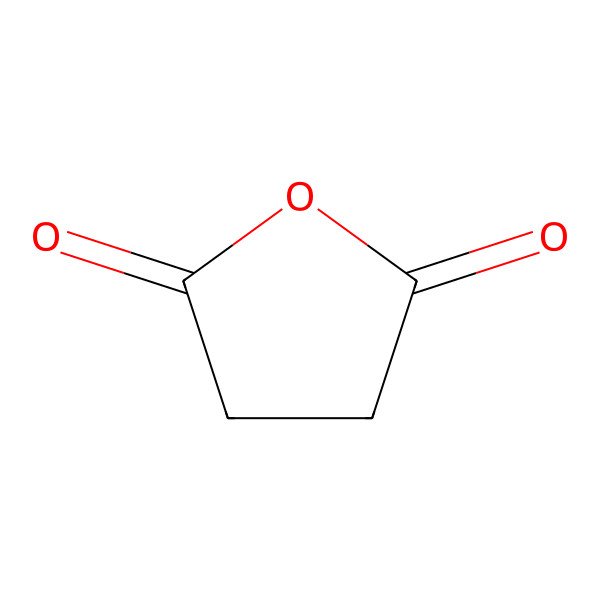
| 108-30-5 |
| dihydrofuran-2,5-dione |
| oxolane-2,5-dione |
| Succinic acid anhydride |
| Succinyl oxide |
| Dihydro-2,5-furandione |
| 2,5-Diketotetrahydrofuran |
| Succinyl anhydride |
| Butanedioic anhydride |
| Rikacid SA |
| 2,5-Furandione, dihydro- |
| Tetrahydro-2,5-dioxofuran |
| 2,5-Dioxotetrahydrofuran |
| Tetrahydro-2,5-furandione |
| Bernsteinsaure-anhydrid |
| NCI-C55696 |
| Succinyloxide |
| Dihydro-furan-2,5-dione |
| NSC 8518 |
| CCRIS 2386 |
| HSDB 792 |
| Dihydro-2,5-diketotetrahydrofuran |
| EINECS 203-570-0 |
| UNII-6RF4O17Z8J |
| MFCD00005525 |
| BRN 0108441 |
| 6RF4O17Z8J |
| Bernsteinsaeureanhydrid |
| DTXSID7021287 |
| Bernsteinsaure-anhydrid [German] |
| CHEBI:36595 |
| AI3-52664 |
| SuccinicAnhydride-13C4 |
| NSC-8518 |
| 2,5(3H,4H)-Furandione |
| Succinic anhydride treated BSA |
| DTXCID701287 |
| EC 203-570-0 |
| 5-17-11-00006 (Beilstein Handbook Reference) |
| EINECS 270-161-1 |
| 68412-02-2 |
| SUCCINIC ANHYDRIDE (IARC) |
| SUCCINIC ANHYDRIDE [IARC] |
| SAA |
| tetrahydrofuran-2,5-dione |
| Anhydride succinique |
| 2, dihydro- |
| Oxolan-2,5-dione |
| WLN: T5VOVTJ |
| butanedioic acid anhydride |
| Dihydro-2, 5-furandione |
| Succinic anhydride, 97% |
| Succinic Anhydride (SAN) |
| 2,5-Furandiona, dihidro- |
| BERNSTEINSAUREANHYDRID |
| tetrahydro-furan-2,5-dione |
| Tetrahydro-2, 5-dioxofuran |
| SCHEMBL16123 |
| MLS002454448 |
| SUCCINIC ANHYDRIDE [MI] |
| CHEMBL1370164 |
| NSC8518 |
| SUCCINIC ANHYDRIDE [HSDB] |
| SUCCINIC ANHYDRIDE [INCI] |
| HMS3039B10 |
| STR01303 |
| Succinic anhydride, >=99% (GC) |
| Tox21_201179 |
| LS-457 |
| STL194303 |
| Succinyl Oxide, Butanedoic Anhydride |
| AKOS000121191 |
| Succinic anhydride, >=97.0% (NT) |
| NCGC00091291-01 |
| NCGC00091291-02 |
| NCGC00258731-01 |
| BP-13070 |
| CAS-108-30-5 |
| HY-79369 |
| SMR001372026 |
| Butanedioic acid,anhydride succinic anhydride |
| CS-0011581 |
| FT-0637061 |
| FT-0652573 |
| S0107 |
| EN300-18021 |
| C19524 |
| D78120 |
| Succinic anhydride, puriss., >=99.0% (NT) |
| Succinic anhydride, SAJ first grade, >=99.0% |
| A801840 |
| Q417847 |
| J-002088 |
| BUTANEDIOIC ACID,ANHYDRIDE SUCCINIC ANHYDRIDE |
| Z57127506 |
| F1908-0090 |
| InChI=1/C4H4O3/c5-3-1-2-4(6)7-3/h1-2H |
| Tetrahydro-2,5-furandione(National Library of Medicine, SIS; ChemIDplus Record for Succinic anhydride (108-30-5) Available from, as of July 21, 2005: http://chem.sis.nlm.nih.gov/chemidplus/chemidlite.jsp) |
|
There are more than 10 synonyms. If you wish to see them all click here.
|