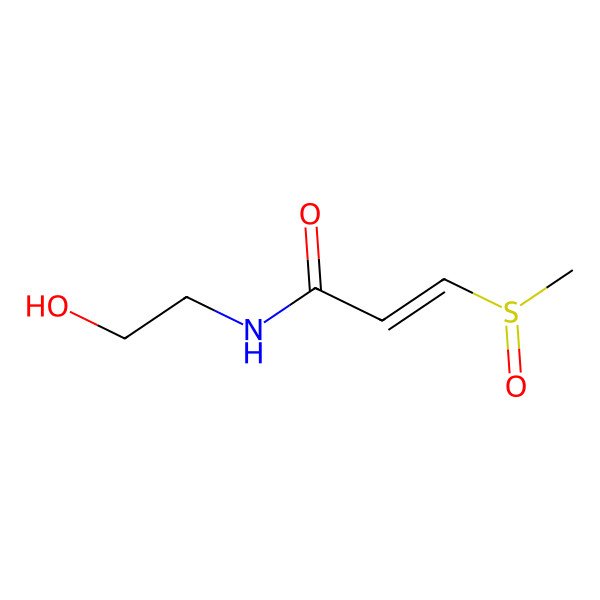
(r)-(+)-trans-N-(2-hydroxyethyl)-3-methylsulphinylpropenamide
| Internal ID | 86e70f2c-9fa2-4041-8d20-55f6c3dda467 |
| Taxonomy | Organic nitrogen compounds > Organonitrogen compounds > Amines > Alkanolamines > 1,2-aminoalcohols > N-acylethanolamines |
| IUPAC Name | (E)-N-(2-hydroxyethyl)-3-[(R)-methylsulfinyl]prop-2-enamide |
| SMILES (Canonical) | CS(=O)C=CC(=O)NCCO |
| SMILES (Isomeric) | C[S@@](=O)/C=C/C(=O)NCCO |
| InChI | InChI=1S/C6H11NO3S/c1-11(10)5-2-6(9)7-3-4-8/h2,5,8H,3-4H2,1H3,(H,7,9)/b5-2+/t11-/m1/s1 |
| InChI Key | LBGAGBWSXXFWGZ-XGQHYKRYSA-N |
| Popularity | 1 reference in papers |
| Molecular Formula | C6H11NO3S |
| Molecular Weight | 177.22 g/mol |
| Exact Mass | 177.04596439 g/mol |
| Topological Polar Surface Area (TPSA) | 85.60 Ų |
| XlogP | -1.80 |
| There are no found synonyms. |
| Target | Value | Probability (raw) | Probability (%) |
|---|---|---|---|
| No predicted properties yet! | |||
Proven Targets:
| CHEMBL ID | UniProt ID | Name | Min activity | Assay type | Source |
|---|---|---|---|---|---|
| No proven targets yet! | |||||
Predicted Targets (via Super-PRED):
| CHEMBL ID | UniProt ID | Name | Probability | Model accuracy |
|---|---|---|---|---|
| CHEMBL4040 | P28482 | MAP kinase ERK2 | 95.56% | 83.82% |
| CHEMBL2581 | P07339 | Cathepsin D | 89.73% | 98.95% |
| CHEMBL3251 | P19838 | Nuclear factor NF-kappa-B p105 subunit | 89.63% | 96.09% |
| CHEMBL5619 | P27695 | DNA-(apurinic or apyrimidinic site) lyase | 84.86% | 91.11% |
| CHEMBL3060 | Q9Y345 | Glycine transporter 2 | 83.68% | 99.17% |
| CHEMBL3892 | Q99500 | Sphingosine 1-phosphate receptor Edg-3 | 80.94% | 97.29% |
| CHEMBL2111367 | P27986 | PI3-kinase p110-alpha/p85-alpha | 80.64% | 94.33% |
Below are displayed all the plants proven (via scientific papers) to contain this
compound!
To see more specific details click the taxa you are interested in.
To see more specific details click the taxa you are interested in.
| Clinacanthus nutans |
| PubChem | 14237624 |
| LOTUS | LTS0039270 |
| wikiData | Q105149245 |