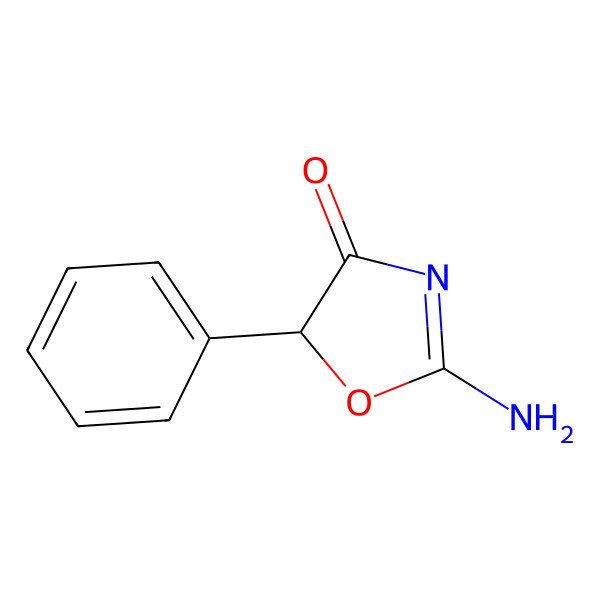
| Phenoxazole |
| Phenylisohydantoin |
| Azoksodon |
| Fenoxazol |
| Azoxodon |
| Dantromin |
| Cylert |
| Pemolin |
| Tradon |
| Azoxodone |
| Centramin |
| Deltamine |
| 2152-34-3 |
| Hyton |
| Nitan |
| Kethamed |
| Sigmadyn |
| Stimulol |
| Okodon |
| Pioxol |
| Pondex |
| Sistra |
| Stimul |
| Tradone |
| Volital |
| Ronyl |
| Phenylpseudohydantoin |
| Pheniminooxazolidinone |
| Betanamin |
| Deltamin |
| Myamin |
| Constimol |
| Endolin |
| Phenalone |
| Phenilone |
| Pomoline |
| Volitol |
| Notair |
| Hyton asa |
| Juston-Wirkstoff |
| Pemolina |
| Senior |
| Sistral |
| Sofro |
| 2-Imino-5-phenyl-4-oxazolidinone |
| Pemolinum |
| Yh 1 |
| 4(5H)-Oxazolone, 2-amino-5-phenyl- |
| NPL 1 |
| 2-Amino-5-phenyl-4(5H)-oxazolone |
| Abbott 13397 |
| 5-Phenylisohydantion |
| 5-Phenyl-2-imino-4-oxazolidinone |
| 5-Phenyl-2-imino-4-oxooxazolidine |
| 2-Amino-5-phenyl-1,3-oxazol-4(5H)-one |
| NSC-25159 |
| 4-Oxazolidinone, 2-imino-5-phenyl- |
| 2-amino-5-phenyl-1,3-oxazol-4-one |
| CHEBI:7953 |
| Pn/135 |
| FWH-352 |
| 2-Imino-4-keto-5-phenyltetrahydrooxazole |
| LA 956 |
| PT 360 |
| NSC-169499 |
| YH-1 |
| 101053-01-4 |
| 2-Amino-5-phenyl-oxazol-4-one |
| H 310 |
| CS 293 |
| LA-956 |
| PIO |
| A 13397 |
| 2-Oxazolin-4-one, 2-amino-5-phenyl- |
| 2-amino-5-phenyl-4-oxazolone |
| 7GAQ2332NK |
| DTXCID503427 |
| Fio |
| DTXSID3023427 |
| C- 293 |
| H 3104 |
| NSC25159 |
| 2-amino-5-phenyl-2-oxazolin-4-one |
| NSC169499 |
| 5-PHENYL-2-IMINOOXAZOLIDIN-4-ONE |
| 2-IMINO-4-OXO-5-PHENYLOXAZOLIDINE |
| PEMOLINE (MART.) |
| PEMOLINE [MART.] |
| WLN: T5OYMV EHJ BUM ER |
| 5-Phenyl-2-imino-4-oxazolidine |
| P 10 |
| SMR000238142 |
| CAS-2152-34-3 |
| 2-Amino-5-phenyl4(5H)-oxazolone |
| Cylert (TN) |
| PEMOLINE [HSDB] |
| PEMOLINE [USAN] |
| PEMOLINE [INN] |
| PEMOLINE [JAN] |
| PEMOLINE [MI] |
| PEMOLINE [VANDF] |
| PEMOLINE [WHO-DD] |
| CHEMBL1177 |
| SCHEMBL41636 |
| Pemoline (JAN/USAN/INN) |
| MLS000759491 |
| MLS001424026 |
| PEMOLINE [ORANGE BOOK] |
| Pemoline, >=98% (HPLC) |
| NRNCYVBFPDDJNE-UHFFFAOYSA- |
| N06BA05 |
| HMS2051C08 |
| HMS3393C08 |
| 5-Phenyl-2-imino-4-oxo-oxazolidin |
| Tox21_112509 |
| BDBM50248019 |
| AKOS005065730 |
| AKOS015888248 |
| Tox21_112509_1 |
| 2-azanyl-5-phenyl-1,3-oxazol-4-one |
| CCG-100848 |
| DB01230 |
| NC00098 |
| NCGC00246967-01 |
| NCGC00246967-02 |
| AC-22513 |
| AS-17043 |
| C-293 |
| FT-0655727 |
| P0392 |
| 2-Amino-5-phenyl-1,3-oxazol-4(5H)-one # |
| C07899 |
| D00744 |
| A800319 |
| A815446 |
| W-107545 |
| InChI=1/C9H8N2O2/c10-9-11-8(12)7(13-9)6-4-2-1-3-5-6/h1-5,7H,(H2,10,11,12) |
|
There are more than 10 synonyms. If you wish to see them all click here.
|