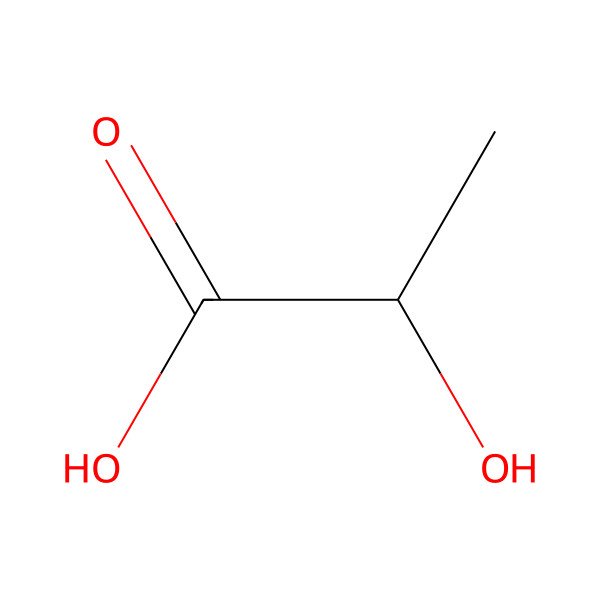
| 79-33-4 |
| L-(+)-Lactic acid |
| (S)-Lactic acid |
| (S)-2-Hydroxypropanoic acid |
| Sarcolactic acid |
| (2S)-2-hydroxypropanoic acid |
| (S)-2-Hydroxypropionic acid |
| (+)-Lactic acid |
| (S)-(+)-Lactic acid |
| Paralactic acid |
| L(+)-LACTIC ACID |
| Lactic acid, L- |
| Tisulac |
| PURAC |
| (S)-lactate |
| Paramilchsaeure |
| L-lactate |
| Propanoic acid, 2-hydroxy-, (2S)- |
| Fleischmilchsaeure |
| (S)-Milchsaeure |
| Acidum sarcolacticum |
| Sarcolacticum acidum |
| L-(+)-alpha-Hydroxypropionic acid |
| Pleo sanvis |
| PH 90 |
| (S)-2-Hydroxypropionsaeure |
| PLLA |
| PROPANOIC ACID, 2-HYDROXY-, (S)- |
| EINECS 201-196-2 |
| UNII-F9S9FFU82N |
| BRN 1720251 |
| F9S9FFU82N |
| CHEBI:422 |
| L( )-2-Hydroxypropionsaeure |
| L(+)-2-Hydroxypropionsaeure |
| (+)-Lactate |
| EC 201-196-2 |
| 4-03-00-00633 (Beilstein Handbook Reference) |
| MFCD00064266 |
| 2-hydroxy-propionic acid |
| 1-Hydroxyethane 1-carboxylic acid |
| Atrisorb |
| Sculptra |
| Ticron |
| (C3-H6-O3)x- |
| L-lacticacid |
| L-Milchsaeure |
| (alpha)-Lactate |
| L-Iactic acid |
| New-Fill |
| Lactic Acid FCC |
| a-Hydroxypropanoate |
| a-Hydroxypropionate |
| 2OP |
| L-Lactic acid (2-hydroxy propionic acid) |
| nchembio867-comp9 |
| (alpha)-Lactic acid |
| alpha-Hydroxypropanoate |
| alpha-Hydroxypropionate |
| L-2-Hydroxypropanoate |
| a-Hydroxypropanoic acid |
| a-Hydroxypropionic acid |
| L-(+) Lactic Acid |
| L-Lactic Acid, 90% |
| (S)-2-Hydroxypropanoate |
| (S)-2-Hydroxypropionate |
| 1-Hydroxyethanecarboxylate |
| L-Lactic acid, anhydrous |
| L-2-Hydroxypropanoic acid |
| bmse000208 |
| bmse000818 |
| bmse000979 |
| D06FZS |
| (S)-2-hydroxy-Propanoate |
| UNII-W92NDP347A |
| L-LACTIC ACID [MI] |
| L-LACTIC ACID [JAN] |
| L-(+)-Lactic acid solution |
| 1-Hydroxyethane 1-carboxylate |
| (S)-2-hydroxy-Propanoic acid |
| (S)-2-hydroxy-propionic acid |
| CHEMBL330546 |
| GTPL2932 |
| L- LACTIC ACID (+) |
| (S)-(+)-2-Hydroxypropanoate |
| L-(+)-Lactic acid, 80% |
| (S)(+)2 hydroxypropionic acid |
| DTXSID6034689 |
| LACTIC ACID, L- [II] |
| DEXTROROTATORY LACTIC ACID |
| (s)(+)-2 hydroxypropionic acid |
| SARCOLACTIC ACID [WHO-DD] |
| L-(+)-Lactic acid 95% liquid |
| 80% (w/w) Lactic Acid Solution |
| L-(+)-Lactic acid solution, 1M |
| L-(+)-Lactic acid, >=98% |
| SARCOLACTICUM ACIDUM [HPUS] |
| (S)-(+)-2-Hydroxypropanoic acid |
| 2-Hydroxypropanoic acid, (S)- # |
| 26811-96-1 |
| HY-Y0479 |
| DL6049 |
| s6250 |
| AKOS025146504 |
| DB14475 |
| L-Lactic acid, crystalline, 98.0%+ |
| (S)-LACTIC ACID [EP MONOGRAPH] |
| L-(+)-Lactic acid, analytical standard |
| LS-87453 |
| CS-0015266 |
| FT-0773848 |
| L0165 |
| EN300-91905 |
| C00186 |
| D71144 |
| L-0990 |
| L-1000 |
| L-(+)-Lactic acid, BioXtra, >=98% (titration) |
| L-(+)-Lactic acid, Vetec(TM) reagent grade, 86% |
| Q27080955 |
| 5E39D33D-2F71-4C24-BC7A-5E6F27E4CF83 |
| L+Lactic Acid, Free Acid (S)-2-Hydroxypropionic acid, Sarcolactic acid |
|
There are more than 10 synonyms. If you wish to see them all click here.
|