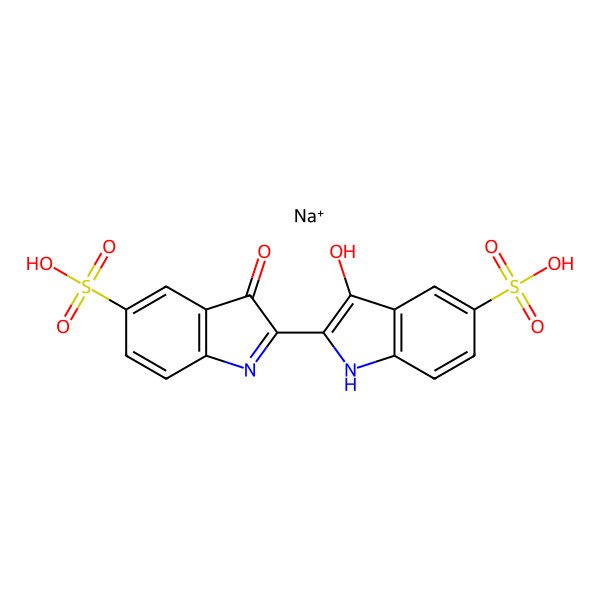
| Indigotine |
| Murabba |
| Indigotine Lake |
| Soluble Indigo |
| Indocarmine F |
| Indigotine B |
| Indigotine I |
| Indigotine N |
| Intense Blue |
| Indigotin I |
| Indigotine IA |
| Cilefa Blue R |
| Indigo Carmine A |
| Indigo Carmine X |
| Grape Blue A |
| Airedale Blue IN |
| Acid Blue W |
| HD Indigo Carmine |
| Indigo Carmine AC |
| Indigo Carmine BP |
| Food Blue 2 |
| Indigotine Blue LZ |
| Atul Indigo Carmine |
| Soluble Indigo Blue |
| Bucacid indigotine B |
| Grape Blue A Geigy |
| Maple Indigo Carmine |
| Edicol Supra Blue X |
| Amacid Brilliant Blue |
| Indigo Carmine Powder |
| Mitsui Indigo Carmine |
| FD and C Blue 2 |
| Aniline Carmine powder |
| Acid Leather Blue IC |
| Dolkwal Indigo Carmine |
| San-ei Indigo Carmine |
| Usacert Blue No. 2 |
| Canacert Indigo Carmine |
| Hexacert Blue No. 2 |
| Indigotine disodium salt |
| Indigotine Conc. Powder |
| Indigotine Extra Pure A |
| C.I. Natural Blue 2 |
| HD Indigo Carmine Supra |
| Indigo Carmine Conc. FQ |
| A.F. Blue No. 2 |
| Sodium indigotindisulfonate |
| 1311 Blue |
| L Blue Z 5010 |
| 12070 Blue |
| Hexacol Indigo Carmine Supra |
| Indigo Carmine Disodium Salt |
| Carmine Blue (biological stain) |
| Disodium indigo-5,5-disulfonate |
| NSC8646 |
| Sodium 5,5'-indigotindisulfonate |
| Indigo Carmine (Biological Stain) |
| C.I. Food Blue 1, disodium salt |
| Disodium 5,5'-indigotin disulfonate |
| Indigo disulfonate (Biological Stain) |
| Disodium salt of 1-indigotin-S,S'-disulfonic acid |
| FD&C Blue 2 |
| Sodium 5,5'-indigodisulfonate |
| Disodium 5,5'-indigodisulfonate |
| WLN: T56 BMYVJ GSWO CU- CT56 BMYVJ GSWO &-NA- 2 |
| [.DELTA.2,5'-disulfonic acid, 3,3'-dioxo-, disodium salt |
| [.delta.(sup2,5'-disulfonic acid, 3,3'-dioxo-, disodium salt |
| 1H-Indole-5-sulfonic acid,3-dihydro-3-oxo-5-sulfo-2H-indol-2-ylidene)-2,3-dihydro-3-oxo-, disodium salt |
| 1H-Indole-5-sulfonic acid,3-dihydro-3-oxo-5-sulfo-2H-indol-2-ylidine)-2,3-dihydro-3-oxo-, disodium salt |
|
There are more than 10 synonyms. If you wish to see them all click here.
|