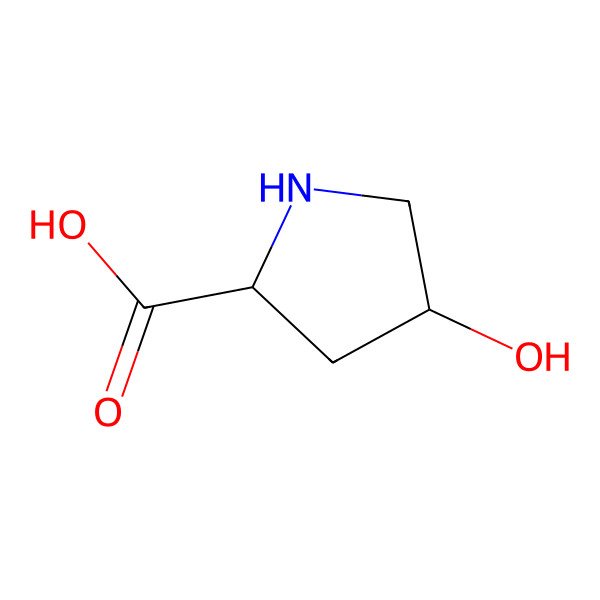
| hydroxyproline |
| 51-35-4 |
| L-Hydroxyproline |
| (2S,4R)-4-Hydroxypyrrolidine-2-carboxylic acid |
| trans-4-Hydroxyproline |
| L-4-hydroxyproline |
| Hypro |
| H-Hyp-OH |
| hydroxy-L-proline |
| 4-Hydroxy-L-proline |
| 4-hydroxyproline |
| trans-L-Hydroxyproline |
| L-Proline, 4-hydroxy-, (4R)- |
| trans-Hydroxyproline |
| (4R)-4-hydroxy-L-proline |
| 4-Hydroxy-2-pyrrolidinecarboxylic acid |
| delta-Hydroxyproline |
| L-Proline, 4-hydroxy-, trans- |
| Proline, 4-hydroxy-, L- |
| Hydroxyproline (VAN) |
| L-Proline, 4-hydroxy- |
| Ls-Hydroxyproline |
| UNII-RMB44WO89X |
| RMB44WO89X |
| (2S,4R)-4-hydroxyproline |
| 4-L-Hydroxyproline |
| Proline, 4-hydroxy- (VAN) |
| LUMISTOR |
| CHEBI:18095 |
| (2S,4R)-trans-4-hydroxyproline |
| trans-4-hydroxy-proline |
| L-threo-4-hydroxyproline |
| EINECS 200-091-9 |
| NSC 46704 |
| NSC-46704 |
| Proline, 4-hydroxy- |
| CHEMBL352418 |
| Hyp |
| trans-L-4-hydroxyproline |
| DTXSID10883225 |
| (2S,4R)-4-hydroxy-2-pyrrolidinecarboxylic acid |
| NSC46704 |
| trans-oxyproline |
| 618-28-0 |
| trans-hydroxy-L-proline |
| (2S,4R)-rel-4-Hydroxypyrrolidine-2-carboxylic acid |
| L-Hydroxyprolin |
| trans-L-4-hydroxy-proline |
| MFCD00064320 |
| L-Proline, 4-hydroxy-, trans- (9CI) |
| SMR000857104 |
| 6912-67-0 |
| CHEBI:18240 |
| hydroxiproline |
| oxyproline |
| hydroxy-proline |
| L-Hydroxyproline; |
| H-trans-Hyp-OH |
| Hydroxyproline,(l) |
| L-Proline, trans- |
| NMH-Pro |
| (4r)-hydroxyproline |
| 4(R)-hydroxyproline |
| (2S,4R)-(-)-4-Hydroxy-2-Pyrrolidinecarboxylic Acid |
| .delta.-Hydroxyproline |
| L-Hydroxyproline,(S) |
| L- 4- hydroxyproline |
| L-4-transhydroxyproline |
| 4 HYDROXYPROLINE |
| trans-4-hydroxyL-proline |
| trans-4-D-Hydroxyproline |
| Trans4-hydroxy-L-proline |
| bmse000123 |
| bmse000966 |
| D02YMI |
| L-trans-4-hydroxy-proline |
| trans-4-hydroxy(L)proline |
| HYDROXYPROLINE [MI] |
| trans-4-hydroxyl-l-proline |
| SCHEMBL21185 |
| HYDROXYPROLINE [INCI] |
| MLS001332463 |
| MLS001332464 |
| trans-4-hydroxy-(L)-proline |
| (2S, 4R)-4-hydroxyproline |
| GTPL4704 |
| (2s,4r)-4-hydroxy-l-proline |
| HYDROXYPROLINE [WHO-DD] |
| (2S, 4R)-4-hydroxy-proline |
| CHEBI:24741 |
| PMMYEEVYMWASQN-DMTCNVIQSA-N |
| DTXCID001022774 |
| L-prolina, 4-hidroxi-, (4R)- |
| BDBM50357233 |
| s5820 |
| trans-4-Hydroxy-L-proline, >=99% |
| AKOS007930607 |
| AC-2249 |
| AM81829 |
| CS-W008928 |
| DB08847 |
| PS-5807 |
| (2S,4R)-2-carboxy-4-hydroxypyrrolidine |
| BP-10638 |
| HY-40135 |
| NCI60_004102 |
| L-Hydroxyproline;trans-4-Hydroxy-L-Proline |
| 4(R)-hydroxy-2(S)-pyrrolidinecarboxylic acid |
| EN300-53732 |
| C01157 |
| H-7290 |
| M03214 |
| P16433 |
| trans-4-Hydroxy-L-proline, analytical standard |
| (2S, 4R)-4-hydroxy-2-pyrrolidinecarboxylic acid |
| (2S,4R)-4-hydroxy-pyrrolidine-2-carboxylic acid |
| 4-(R)-hydroxy-pyrrolidine-2-(S)-carboxylic acid |
| A905519 |
| (2S, 4R)-4-hydroxy-2-pyrrolidine-carboxylic acid |
| J-525022 |
| Q27089020 |
| trans-4-Hydroxy-L-proline, BioXtra, >=99.0% (NT) |
| F8889-8652 |
| Z802856442 |
| F05487BD-FA1D-4C80-83AD-3A91E2078031 |
| trans-4-Hydroxy-L-proline, microbial, cell culture tested |
| trans-4-Hydroxy-L-proline, Vetec(TM) reagent grade, 99% |
| trans-4-Hydroxy-L-proline, BioReagent, suitable for cell culture, >=98.5% |
| Hydroxyproline, Pharmaceutical Secondary Standard; Certified Reference Material |
|
There are more than 10 synonyms. If you wish to see them all click here.
|