| 93479-97-1 |
| Amaryl |
| Amarel |
| Glimepirid |
| Glimepirida |
| Glimepiridum |
| cis-Glimepiride |
| Endial |
| Hoe-490 |
| 684286-46-2 |
| Glimepride |
| HOE 490 |
| Roname |
| Glimepiridum [Latin] |
| Glimepirida [Spanish] |
| Glimepiride, cis- |
| GLIMPERIDE |
| Glimepiride Impurity A |
| Glymepirid |
| Glorion |
| Glemax |
| Glimer |
| Solosa |
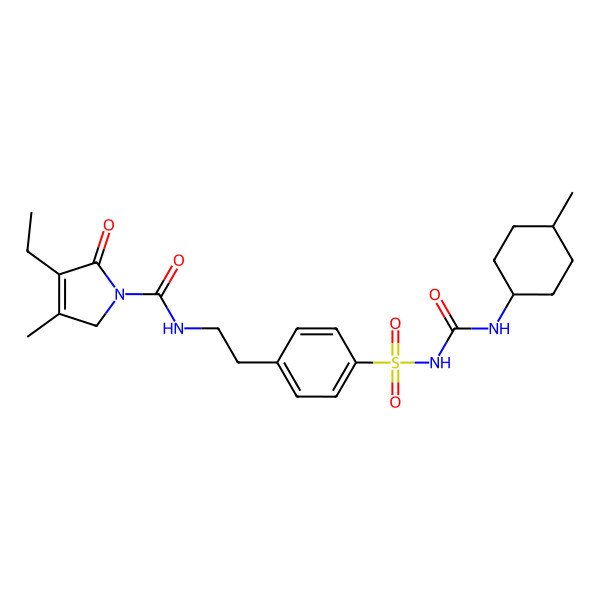
| 4-ethyl-3-methyl-N-[2-[4-[(4-methylcyclohexyl)carbamoylsulfamoyl]phenyl]ethyl]-5-oxo-2H-pyrrole-1-carboxamide |
| Amaryl (TN) |
| Glimepiride [USAN:BAN:INN] |
| CCRIS 7083 |
| CHEBI:5383 |
| 1-((p-(2-(3-Ethyl-4-methyl-2-oxo-3-pyrroline-1-carboxamido)ethyl)phenyl)sulfonyl)-3-(trans-4-methylcyclohexyl)urea |
| C24H34N4O5S |
| UNII-24T6XIR2MZ |
| NSC-759809 |
| BRN 5365754 |
| diameprid |
| glimax |
| glirid |
| 24T6XIR2MZ |
| HOE490 |
| 261361-60-8 |
| DTXSID5040675 |
| UNII-6KY687524K |
| Glimepiride [USAN:USP:INN:BAN] |
| 6KY687524K |
| NCGC00016960-03 |
| CAS-93479-97-1 |
| Glista OD |
| 1H-Pyrrole-1-carboxamide, 3-ethyl-2,5-dihydro-4-methyl-N-(2-(4-(((((4-methylcyclohexyl)amino)carbonyl)amino)sulfonyl)phenyl)ethyl)-2-oxo-, trans- |
| 3-ethyl-4-methyl-N-[2-(4-{[(trans-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamide |
| DTXCID3020675 |
| Novo-glimepiride |
| PMS-glimepiride |
| Ratio-glimepiride |
| Sandoz glimepiride |
| 1-{[4-(2-{[(3-ethyl-4-methyl-2-oxo-2,5-dihydro-1H-pyrrol-1-yl)carbonyl]amino}ethyl)phenyl]sulfonyl}-3-(trans-4-methylcyclohexyl)urea |
| 1H-Pyrrole-1-carboxamide, 2,5-dihydro-3-ethyl-4-methyl-N-(2-(4-(((((4-methylcyclohexyl)amino)carbonyl)amino)sulfonyl)phenyl)ethyl)-2-oxo-, trans- |
| 3-ethyl-4-methyl-n-(4-(n-((1r,4r)-4-methylcyclohexylcarbamoyl)sulfamoyl)phenethyl)-2-oxo-2,5-dihydro |
| 3-ethyl-4-methyl-N-{2-[4-({[(4-methylcyclohexyl)carbamoyl]amino}sulfonyl)phenyl]ethyl}-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamide |
| SMR000466368 |
| SR-05000001508 |
| gimepiride |
| vitamine |
| Niddaryl |
| Sugral |
| Glimepiride,(S) |
| 1-[[4-[2-[[(3-Ethyl-4-methyl-2-oxo-2,3-dihydro-1H-pyrrol-1-yl)carbonyl]amino]ethyl]phenyl]sulphonyl]-3-(cis-4-methylcyclohexyl)urea (cis-Glimepiride) |
| 3-ethyl-4-methyl-N-[2-[4-[(4-methylcyclohexyl)carbamoylsulfamoyl]phenyl]ethyl]-2-oxo-5H-pyrrole-1-carboxamide |
| Amaryl, Glista OD |
| Glimepiride- Bio-X |
| MFCD00878417 |
| Glimepirida Parke-Davis |
| CPD000466368 |
| GLIMEPIRIDE [MI] |
| Prestwick0_000651 |
| Prestwick1_000651 |
| Prestwick2_000651 |
| Prestwick3_000651 |
| GLIMEPIRIDE [INN] |
| GLIMEPIRIDE [JAN] |
| GLIMEPIRIDE [USAN] |
| D0B2GI |
| GLIMEPIRIDE [VANDF] |
| CHEMBL1481 |
| GLIMEPIRIDE [MART.] |
| Oprea1_382896 |
| SCHEMBL16084 |
| SCHEMBL16086 |
| BSPBio_000681 |
| GLIMEPIRIDE [USP-RS] |
| GLIMEPIRIDE [WHO-DD] |
| GLIMEPIRIDE CIS-ISOMER |
| MLS000759495 |
| MLS001076674 |
| MLS001401419 |
| MLS003915622 |
| MLS006011260 |
| Glimepiride (JAN/USP/INN) |
| SPBio_002602 |
| GLIMEPIRIDE [EMA EPAR] |
| Glimepiride [USAN:INN:BAN] |
| BPBio1_000751 |
| CHEMBL149223 |
| GTPL6820 |
| SCHEMBL8738802 |
| Glimepiride (JP17/USP/INN) |
| Amaryl, Glista OD, Glimepiride |
| SCHEMBL14371714 |
| SCHEMBL14965363 |
| CHEBI:92609 |
| DTXSID20861130 |
| GLIMEPIRIDE [ORANGE BOOK] |
| Glimepiride for system suitability |
| WIGIZIANZCJQQY-RUCARUNLSA-N |
| WIGIZIANZCJQQY-UHFFFAOYSA-N |
| GLIMEPIRIDE [EP MONOGRAPH] |
| GLIMEPIRIDE [USP IMPURITY] |
| HMS1570C03 |
| HMS2052L03 |
| HMS2090K18 |
| HMS2097C03 |
| HMS2235L07 |
| HMS3269A09 |
| HMS3372O07 |
| HMS3394L03 |
| HMS3413K06 |
| HMS3654F17 |
| HMS3677K06 |
| HMS3714C03 |
| Pharmakon1600-01504915 |
| GLIMEPIRIDE [USP MONOGRAPH] |
| BCP05331 |
| DUETACT COMPONENT GLIMEPIRIDE |
| HY-B0104 |
| Tox21_110713 |
| AC-476 |
| BDBM50237590 |
| C24-H34-N4-O5-S |
| NSC759809 |
| NSC813217 |
| s1344 |
| STL451059 |
| STL453194 |
| AKOS015894919 |
| AKOS015969663 |
| Glimepiride, >=98% (HPLC), solid |
| Tox21_110713_1 |
| AB07644 |
| BCP9000728 |
| CCG-101156 |
| CS-1844 |
| CS-O-30930 |
| DB00222 |
| GLIMEPIRIDE COMPONENT OF DUETACT |
| KS-5238 |
| NC00406 |
| NSC 759809 |
| NSC-813217 |
| NCGC00016960-01 |
| NCGC00016960-02 |
| NCGC00016960-04 |
| NCGC00016960-05 |
| NCGC00016960-07 |
| NCGC00161404-01 |
| NCGC00161404-02 |
| NCGC00181757-01 |
| NCGC00371061-02 |
| NCGC00371061-06 |
| BG164507 |
| SMR001550123 |
| Glimepiride 100 microg/mL in Acetonitrile |
| LS-136752 |
| AB00513874 |
| CS-0165191 |
| FT-0626713 |
| FT-0668978 |
| G0395 |
| GLIMEPIRIDE IMPURITY A [EP IMPURITY] |
| SW196369-4 |
| 64598P |
| C07669 |
| D00593 |
| AB00513874-06 |
| AB00513874-08 |
| AB00513874-09 |
| AB00513874_10 |
| AB00513874_11 |
| A844609 |
| A899888 |
| EN300-19873446 |
| Q425027 |
| Q-201158 |
| SR-05000001508-1 |
| SR-05000001508-2 |
| SR-05000001508-3 |
| BRD-K34776109-001-03-4 |
| BRD-K42693031-001-01-8 |
| Q27253874 |
| Z1501475009 |
| Glimepiride, British Pharmacopoeia (BP) Reference Standard |
| Glimepiride, European Pharmacopoeia (EP) Reference Standard |
| 3-ethyl-4-methyl-N-(4-(N-((1r,4r)-4-methylcyclohexylcarbamoyl) |
| Glimepiride, United States Pharmacopeia (USP) Reference Standard |
| sulfamoyl)phenethyl)-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamide |
| Glimepiride for system suitability, European Pharmacopoeia (EP) Reference Standard |
| ;3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car |
| ;cis-3-Ethyl-2,5-dihydro-4-Methyl-N-[2-[4-[[[[(cis-4-Methylcyclohexyl)aMino]carbonyl]aMino]sulfonyl]phenyl]ethyl]-2-oxo-1H-pyrrole-1-carboxaMide |
| 1-((4-(2-(((3-ETHYL-4-METHYL-2-OXO-2,3-DIHYDRO-1H-PYRROL-1-YL)CARBONYL)AMINO)ETHYL)PHENYL)SULFONYL)-3-(CIS-4-METHYLCYCLOHEXYL)UREA |
| 1H-PYRROLE-1-CARBOXAMIDE, 3-ETHYL-2,5-DIHYDRO-4-METHYL-N-(2-(4-(((((CIS-4-METHYLCYCLOHEXYL)AMINO)CARBONYL)AMINO)SULFONYL)PHENYL)ETHYL)-2-OXO- |
| 1H-Pyrrole-1-carboxamide, 3-ethyl-2,5-dihydro-4-methyl-N-[2-[4-[[[[(trans- 4-methylcyclohexyl)amino]carbonyl]amino]sulfonyl]phenyl]ethyl]-2-oxo- |
| 1H-Pyrrole-1-carboxamide, 3-ethyl-2,5-dihydro-4-methyl-N-[2-[4-[[[[(trans-4-methylcyclohexyl)amino]carbonyl]amino]sulfonyl]phenyl]ethyl]-2-oxo- |
| 3-Ethyl-2,5-dihydro-4-methyl-N-[2-[4-[[[[(4-methylcyclohexyl)amino]carbonyl]amino]sulfonyl]phenyl]ethyl]-2-oxo-1H-pyrrole-1-carboxamide |
| 3-Ethyl-2,5-dihydro-4-methyl-N-[2-[4-[[[[(trans-4-methylcyclohexyl)amino]carbonyl]amino]sulfonyl]phenyl]ethyl]-2-oxo-1H-pyrrole-1-carboxamide |
| 3-ethyl-4-methyl-2-oxo-N-(2-{4-[({[(1r,4r)-4-methylcyclohexyl]carbamoyl}amino)sulfonyl]phenyl}ethyl)-2,5-dihydro-1H-pyrrole-1-carboxamide |
| 3-ethyl-4-methyl-N-(4-(N-(((1r,4r)-4-methylcyclohexyl)carbamoyl)sulfamoyl)phenethyl)-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamide |
| 3-ethyl-4-methyl-N-(4-(N-(((1s,4s)-4-methylcyclohexyl)carbamoyl)sulfamoyl)phenethyl)-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamide? (Glimepiride Impurity pound(c) |
| 3-ETHYL-4-METHYL-N-(4-(N-((1R,4R)-4-METHYLCYCLOHEXYLCARBAMOYL)SULFAMOYL)PHENETHYL)-2-OXO-2,5-DIHYDRO-1H-PYRROLE-1-CARBOXAMIDE |
| 3-Ethyl-4-methyl-N-(4-(N-((rel-(1r,4r)-4-methylcyclohexyl)carbamoyl)sulfamoyl)phenethyl)-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamide |
| 3-ethyl-4-methyl-N-(4-(N-((trans-4-methylcyclohexyl)carbamoyl)sulfamoyl)phenethyl)-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamide |
| 3-ethyl-4-methyl-N-[2-(4-{[(4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamide |
| 3-Ethyl-4-Methyl-N-[2-(4-{[(Cis-4-Methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-Oxo-2,5-Dihydro-1h-Pyrrole-1-Carboxamide |
| 3-Ethyl-4-methyl-N-[2-[4-[[3-(4-methylcyclohexyl)ureido]sulfonyl]phenyl]ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamide |
| N'-{[4-(2-{[(3-ethyl-4-methyl-2-oxo-2,5-dihydro-1H-pyrrol-1-yl)carbonyl]amino}ethyl)phenyl]sulfonyl}-N-(4-methylcyclohexyl)carbamimidic acid |
| N-(4-ethyl-3-methyl-5-oxo-2H-pyrrol-1-yl)-3-[4-[(4-methylcyclohexyl)carbamoylsulfamoyl]phenyl]propanamide;GLIMEPIRIDE |
| N-[4-[2-(3-ethyl-4-methyl-2-oxo-3-pyrroline-1-carboxamido)-ethyl]-benzenesulfonyl]-N'-4-methylcyclohexylurea |
| trans-1-(4-(2-(3-ethyl-4-Me-2-oxo-2,5-dihydro-1H-pyrrole-1-carboxamido)ethyl)phenylsulfonyl)-3-(4-methylcyclohexyl)urea |
| trans-3-Ethyl-2,5-dihydro-4-methyl-N-[2-[4-[[[[(4-methyl cyclohexyl)amino]carbonyl]amino]sulfonyl]phenyl]ethyl]-2-oxo-1H-pyrrole-1-carboxamide |
| Trans-3-Ethyl-2,5-dihydro-4-methyl-N-[2-[4-[[[[(4-methylcyclohexyl)amino]carbonyl]amino]sulfonyl]phenyl]ethyl]-2-oxo-1H-pyrrole-1-carboxamide |
| trans-3-Ethyl-2,5-dihydro-4-methyl-N-[2-[4-[[[[trans-4-methylcyclohexyl)amino]carbonyl]amino]sulfonyl]phenyl]ethyl]-2-oxo-1H-pyrrole-1-carboxyamide |
|
There are more than 10 synonyms. If you wish to see them all click here.
|