Fluoxetine
| Internal ID | 5b65d46c-6f75-4df1-ac07-4e6b096ab0ad |
| Taxonomy | Benzenoids > Benzene and substituted derivatives > Trifluoromethylbenzenes |
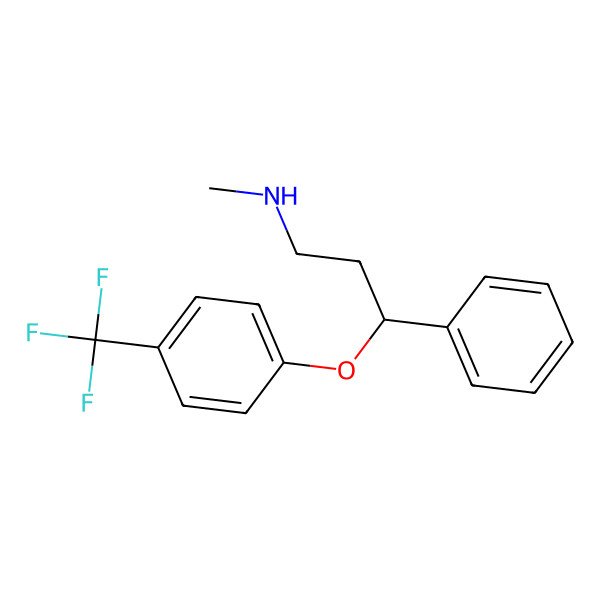
| IUPAC Name | N-methyl-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine |
| SMILES (Canonical) | CNCCC(C1=CC=CC=C1)OC2=CC=C(C=C2)C(F)(F)F |
| SMILES (Isomeric) | CNCCC(C1=CC=CC=C1)OC2=CC=C(C=C2)C(F)(F)F |
| InChI | InChI=1S/C17H18F3NO/c1-21-12-11-16(13-5-3-2-4-6-13)22-15-9-7-14(8-10-15)17(18,19)20/h2-10,16,21H,11-12H2,1H3 |
| InChI Key | RTHCYVBBDHJXIQ-UHFFFAOYSA-N |
| Popularity | 28,676 references in papers |
| Molecular Formula | C17H18F3NO |
| Molecular Weight | 309.33 g/mol |
| Exact Mass | 309.13404868 g/mol |
| Topological Polar Surface Area (TPSA) | 21.30 Ų |
| XlogP | 4.00 |
| Atomic LogP (AlogP) | 4.44 |
| H-Bond Acceptor | 2 |
| H-Bond Donor | 1 |
| Rotatable Bonds | 6 |
| 54910-89-3 |
| Prozac |
| Pulvules |
| Eufor |
| Animex-On |
| Fluoxetina |
| Fluval |
| Portal |
| Fluoxetinum |
| Fluoxetin |
| There are more than 10 synonyms. If you wish to see them all click here. |
| Target | Value | Probability (raw) | Probability (%) |
|---|---|---|---|
| Human Intestinal Absorption | + | 1.0000 | 100.00% |
| Caco-2 | + | 0.9313 | 93.13% |
| Blood Brain Barrier | + | 1.0000 | 100.00% |
| Human oral bioavailability | + | 0.9286 | 92.86% |
| Subcellular localzation | Mitochondria | 0.5725 | 57.25% |
| OATP2B1 inhibitior | - | 1.0000 | 100.00% |
| OATP1B1 inhibitior | + | 0.9291 | 92.91% |
| OATP1B3 inhibitior | + | 0.9480 | 94.80% |
| MATE1 inhibitior | - | 1.0000 | 100.00% |
| OCT2 inhibitior | + | 0.8750 | 87.50% |
| BSEP inhibitior | + | 0.8441 | 84.41% |
| P-glycoprotein inhibitior | - | 0.7443 | 74.43% |
| P-glycoprotein substrate | - | 0.8446 | 84.46% |
| CYP3A4 substrate | + | 0.6340 | 63.40% |
| CYP2C9 substrate | + | 1.0000 | 100.00% |
| CYP2D6 substrate | + | 0.6779 | 67.79% |
| CYP3A4 inhibition | + | 0.7959 | 79.59% |
| CYP2C9 inhibition | - | 0.9070 | 90.70% |
| CYP2C19 inhibition | + | 0.8993 | 89.93% |
| CYP2D6 inhibition | + | 0.8932 | 89.32% |
| CYP1A2 inhibition | + | 0.9107 | 91.07% |
| CYP2C8 inhibition | - | 0.9882 | 98.82% |
| CYP inhibitory promiscuity | + | 0.7149 | 71.49% |
| UGT catelyzed | - | 0.0000 | 0.00% |
| Carcinogenicity (binary) | - | 0.6756 | 67.56% |
| Carcinogenicity (trinary) | Non-required | 0.7114 | 71.14% |
| Eye corrosion | - | 0.9886 | 98.86% |
| Eye irritation | - | 0.9541 | 95.41% |
| Skin irritation | - | 0.6520 | 65.20% |
| Skin corrosion | - | 0.7758 | 77.58% |
| Ames mutagenesis | - | 0.7600 | 76.00% |
| Human Ether-a-go-go-Related Gene inhibition | + | 0.9161 | 91.61% |
| Micronuclear | - | 0.6300 | 63.00% |
| Hepatotoxicity | + | 0.8750 | 87.50% |
| skin sensitisation | - | 0.8401 | 84.01% |
| Respiratory toxicity | + | 0.9667 | 96.67% |
| Reproductive toxicity | + | 0.7444 | 74.44% |
| Mitochondrial toxicity | + | 0.9625 | 96.25% |
| Nephrotoxicity | + | 0.5092 | 50.92% |
| Acute Oral Toxicity (c) | III | 0.7805 | 78.05% |
| Estrogen receptor binding | + | 0.8906 | 89.06% |
| Androgen receptor binding | + | 0.6307 | 63.07% |
| Thyroid receptor binding | + | 0.6494 | 64.94% |
| Glucocorticoid receptor binding | - | 0.7332 | 73.32% |
| Aromatase binding | - | 0.5099 | 50.99% |
| PPAR gamma | + | 0.7852 | 78.52% |
| Honey bee toxicity | - | 0.8641 | 86.41% |
| Biodegradation | - | 0.8750 | 87.50% |
| Crustacea aquatic toxicity | + | 0.7400 | 74.00% |
| Fish aquatic toxicity | + | 0.6474 | 64.74% |
Proven Targets:
| CHEMBL ID | UniProt ID | Name | Min activity | Assay type | Source |
|---|---|---|---|---|---|
| CHEMBL220 | P22303 | Acetylcholinesterase |
130 nM |
IC50 |
PMID: 20053484
|
| CHEMBL1867 | P08913 | Alpha-2a adrenergic receptor |
6.31 nM 6.31 nM |
Ki Ki |
PMID: 11262089
via Super-PRED |
| CHEMBL1942 | P18089 | Alpha-2b adrenergic receptor |
1528 nM |
IC50 |
via CMAUP
|
| CHEMBL3622 | P33261 | Cytochrome P450 2C19 |
40 nM |
IC50 |
PMID: 26355532
|
| CHEMBL289 | P10635 | Cytochrome P450 2D6 |
700 nM |
IC50 |
via CMAUP
|
| CHEMBL238 | Q01959 | Dopamine transporter |
19500 nM 19500 nM 9120.11 nM 18400 nM 6000 nM 18400 nM 6000 nM 4400 nM 4400 nM |
IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 |
PMID: 19256502
PMID: 19014888 PMID: 23602445 PMID: 24974340 PMID: 18667309 PMID: 25221656 PMID: 18550369 PMID: 16750363 PMID: 16750359 |
| CHEMBL240 | Q12809 | HERG |
10 nM 457.09 nM 3100 nM 10 nM 1513.56 nM 1513.56 nM 1513.56 nM |
IC50 IC50 IC50 IC50 IC50 IC50 IC50 |
via Super-PRED
PMID: 21185626 PMID: 20637635 PMID: 23919353 PMID: 18448342 PMID: 12873512 PMID: 15911273 |
| CHEMBL264 | Q9Y5N1 | Histamine H3 receptor |
7300 nM 7300 nM 7300 nM 7300 nM |
Ki Ki Ki Ki |
PMID: 17616397
PMID: 17307358 PMID: 17127059 PMID: 17107798 |
| CHEMBL5514 | P42858 | Huntingtin |
35481.3 nM |
Potency |
via CMAUP
|
| CHEMBL216 | P11229 | Muscarinic acetylcholine receptor M1 |
3203 nM |
IC50 |
via CMAUP
|
| CHEMBL245 | P20309 | Muscarinic acetylcholine receptor M3 |
3595 nM |
IC50 |
via CMAUP
|
| CHEMBL2035 | P08912 | Muscarinic acetylcholine receptor M5 |
1358 nM |
IC50 |
via CMAUP
|
| CHEMBL222 | P23975 | Norepinephrine transporter |
440 nM 5200 nM 5200 nM 1020 nM 1303 nM 2000 nM 2000 nM 4410 nM 563 nM 1020 nM 6309.57 nM 563 nM |
Ki IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 |
via Super-PRED
PMID: 16750363 PMID: 16750359 PMID: 19256502 via CMAUP PMID: 18550369 PMID: 18667309 PMID: 24974340 PMID: 19329313 PMID: 19014888 PMID: 23602445 PMID: 18771916 |
| CHEMBL1293235 | P02545 | Prelamin-A/C |
1412.5 nM |
Potency |
via CMAUP
|
| CHEMBL2842 | P42345 | Serine/threonine-protein kinase mTOR |
26121.6 nM 18492.7 nM 23280.9 nM |
Potency Potency Potency |
via CMAUP
via CMAUP via CMAUP |
| CHEMBL224 | P28223 | Serotonin 2a (5-HT2a) receptor |
710 nM 194 nM |
IC50 IC50 |
via Super-PRED
via CMAUP |
| CHEMBL225 | P28335 | Serotonin 2c (5-HT2c) receptor |
160 nM 119 nM |
IC50 IC50 |
via Super-PRED
via CMAUP |
| CHEMBL3371 | P50406 | Serotonin 6 (5-HT6) receptor |
1661 nM |
IC50 |
via CMAUP
|
| CHEMBL228 | P31645 | Serotonin transporter |
9.4 nM 7.3 nM 9.4 nM 10 nM 9.4 nM 7.2 nM 9.4 nM 9.4 nM 2.5 nM 10 nM 16 nM 9.4 nM 9.4 nM 5.012 nM 16 nM 7 nM 3.1 nM 8.6 nM 9.4 nM 5.2 nM 9.4 nM 5.012 nM 0.72 nM 0.51 nM |
IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 IC50 Ki IC50 |
PMID: 20462211
PMID: 19256502 PMID: 18557608 PMID: 19329313 PMID: 19632110 PMID: 24012181 PMID: 19713106 PMID: 19722525 PMID: 20034793 PMID: 18771916 PMID: 16750359 PMID: 20131864 PMID: 20378347 PMID: 21739935 PMID: 16750363 PMID: 19014888 PMID: 20724153 PMID: 23403082 PMID: 21916421 PMID: 21093273 PMID: 18951020 PMID: 21310612 via Super-PRED via CMAUP |
| CHEMBL287 | Q99720 | Sigma opioid receptor |
851.14 nM 499 nM |
IC50 IC50 |
PMID: 7990111
via CMAUP |
| CHEMBL1940 | Q13936 | Voltage-gated L-type calcium channel alpha-1C subunit |
5400 nM |
IC50 |
via CMAUP
|
Predicted Targets (via Super-PRED):
| CHEMBL ID | UniProt ID | Name | Probability | Model accuracy |
|---|---|---|---|---|
| CHEMBL3251 | P19838 | Nuclear factor NF-kappa-B p105 subunit | 95.07% | 96.09% |
| CHEMBL2581 | P07339 | Cathepsin D | 93.33% | 98.95% |
| CHEMBL221 | P23219 | Cyclooxygenase-1 | 92.38% | 90.17% |
| CHEMBL3060 | Q9Y345 | Glycine transporter 2 | 90.41% | 99.17% |
| CHEMBL2337 | P48067 | Glycine transporter 1 | 89.89% | 95.45% |
| CHEMBL2039 | P27338 | Monoamine oxidase B | 89.86% | 92.51% |
| CHEMBL2035 | P08912 | Muscarinic acetylcholine receptor M5 | 89.07% | 94.62% |
| CHEMBL3108638 | O15164 | Transcription intermediary factor 1-alpha | 88.04% | 95.56% |
| CHEMBL5619 | P27695 | DNA-(apurinic or apyrimidinic site) lyase | 87.72% | 91.11% |
| CHEMBL1293249 | Q13887 | Kruppel-like factor 5 | 86.35% | 86.33% |
| CHEMBL216 | P11229 | Muscarinic acetylcholine receptor M1 | 84.59% | 94.23% |
| CHEMBL3401 | O75469 | Pregnane X receptor | 83.98% | 94.73% |
| CHEMBL1821 | P08173 | Muscarinic acetylcholine receptor M4 | 83.94% | 94.08% |
| CHEMBL4026 | P40763 | Signal transducer and activator of transcription 3 | 83.52% | 82.69% |
| CHEMBL1907 | P15144 | Aminopeptidase N | 83.20% | 93.31% |
| CHEMBL1075094 | Q16236 | Nuclear factor erythroid 2-related factor 2 | 82.54% | 96.00% |
| CHEMBL4422 | O14842 | Free fatty acid receptor 1 | 81.00% | 93.33% |
| CHEMBL2335 | P42785 | Lysosomal Pro-X carboxypeptidase | 80.61% | 100.00% |
| CHEMBL2535 | P11166 | Glucose transporter | 80.58% | 98.75% |
| CHEMBL4481 | P35228 | Nitric oxide synthase, inducible | 80.38% | 94.80% |
Below are displayed all the plants proven (via scientific papers) to contain this
compound!
To see more specific details click the taxa you are interested in.
To see more specific details click the taxa you are interested in.
| Hordeum vulgare |
| Salvia miltiorrhiza |