| 1085-98-9 |
| Dichlofluanide |
| Elvaron |
| Euparen |
| Euparene |
| Oiparen |
| Pecudin |
| Eparen |
| Bayer 47531 |
| Dichlorfluanid |
| Diparen |
| KUE 13032c |
| BAY 47531 |
| KU 13-O32-C |
| Caswell No. 297A |
| Dichlorfluanid [Czech] |
| NSC 218451 |
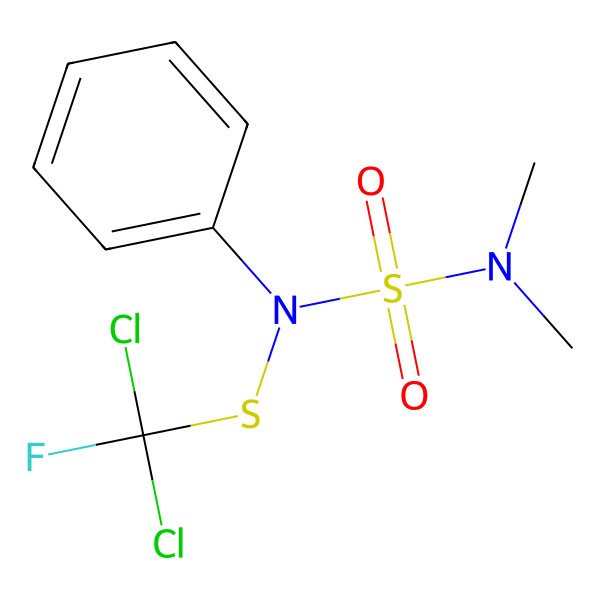
| 1,1-Dichloro-N-((dimethylamino)sulfonyl)-1-fluoro-N-phenylmethanesulfenamide |
| Dichlofluanid [BSI:ISO] |
| Dichlofluanide [ISO-French] |
| N-Dichloroacetyl-N-phenylsemicarbazide |
| CCRIS 3647 |
| Dichlofluanid [ISO] |
| HSDB 1565 |
| EINECS 214-118-7 |
| N-[dichloro(fluoro)methyl]sulfanyl-N-(dimethylsulfamoyl)aniline |
| EPA Pesticide Chemical Code 128844 |
| BRN 2947992 |
| KU 13-032-C |
| UNII-76A92XU36Y |
| N-(Dichlorofluoromethylthio)-N-(dimethylsulfamoyl)-aniline |
| N,N-Dimethyl-N'-phenyl-N'-fluorodichloromethylthiosulfamide |
| DTXSID5041851 |
| CHEBI:74875 |
| N-(Dichlorofluoromethylthio)-N',N'-dimethyl-N-phenylsulfamide |
| 76A92XU36Y |
| Aniline, N-((dichlorofluoromethyl)thio)-N-((dimethylamino)sulfonyl)- |
| N-(Dichlor-fluor-methyl-thio)-N',N'-dimethyl-N-phenyl-schwefel-saeurediamid |
| NSC-218451 |
| N-Dichlorofluoromethylthio-N',N'-dimethyl-N-phenylsulphamide |
| N-Dichlorofluoromethanesulphenyl-N',N'-dimethyl-N-phenylsulphamide |
| Sulfamide, N-((dichlorofluoromethyl)thio)-N',N'-dimethyl-N-phenyl- |
| N-Dichlorfluormethylthio-N',N'-dimethylaminosulfonsaeureanilid [German] |
| 1,1-Dichloro-N-((dimethylamino)sulfonyl)-1-fluoro-N-phenylmethane sulfenamide |
| Methanesulfenamide, 1,1-dichloro-N-((dimethylamino)sulfonyl)-1-fluoro-N-phenyl- |
| Methanesulfenamide, 1,1-dichloro-N-[(dimethylamino)sulfonyl]-1-fluoro-N-phenyl- |
| N-(Dichlor-fluor-methyl-thio)-N',N'-dimethyl-N-phenyl-schwefel-saeurediamid [German] |
| N-Dichlorfluormethylthio-N',N'-dimethylaminosulfonsaeureanilid |
| N-((Dichlorofluoromethyl)thio)-N',N'-dimethyl-N-phenylsulfamide |
| Sulfamide, N-[(dichlorofluoromethyl)thio]-N',N'-dimethyl-N-phenyl- |
| 1,1-Dichloro-N-[(dimethylamino)sulfonyl]-1-fluoro-N-phenylmethanesulfenamide |
| Diclofluanide |
| N,N-Dimethyl-N'-phenyl-N'-((fluorodichloromethyl)thio)sulfamide |
| N,N-Dimethyl-N'-phenyl-N'-[(fluorodichloromethyl)thio]sulfamide |
| N-[(Dichlorofluoromethyl)thio]-N',N'-dimethyl-N-phenylsulfamide |
| Aniline, N-[(dichlorofluoromethyl)thio]-N-[(dimethylamino)sulfonyl]- |
| DICHLOFLUANID [MI] |
| Eparen; Euparen; Euparene |
| DICHLOFLUANID [HSDB] |
| Methanesulfenamide, 1,1-dichloro-N-[(dimethylamino) sulfonyl]-1-fluoro-N-phenyl- |
| SCHEMBL20684 |
| C9H11Cl2FN2O2S2 |
| WLN: GXGFSNR&SWN1&1 |
| CHEMBL1269177 |
| DTXCID3021851 |
| BAYER-47531 |
| Sulfamide,N'-dimethyl-N-phenyl- |
| WURGXGVFSMYFCG-UHFFFAOYSA- |
| n-dichlorofluoromethylthio-n',n'-dimethyl-n-phenylsulfamide |
| WURGXGVFSMYFCG-UHFFFAOYSA-N |
| KUE-13032C |
| BCP28604 |
| Tox21_300960 |
| NSC218451 |
| AKOS015888203 |
| C9-H11-Cl2-F-N2-O2-S2 |
| NCGC00248229-01 |
| NCGC00254862-01 |
| AS-35177 |
| Dichlofluanid 100 microg/mL in Isooctane |
| CAS-1085-98-9 |
| LS-147702 |
| FT-0603081 |
| C18820 |
| Dichlofluanid, PESTANAL(R), analytical standard |
| Q421444 |
| J-002166 |
| N'-Dichlorofluoromethylthio-NN-dimethyl-N'-phenylsulphamide |
| N-(dichloro-fluoromethyl)sulfanyl-N-(dimethylsulfamoyl)aniline |
| N-([Dichloro(fluoro)methyl]sulfanyl)-N',N'-dimethyl-N-phenylsulfamide # |
| N-(Dichlor-fluor-methyl-thio)-N',N'-dimethyl-N-phenylschwefel-saeurediamid |
| Methanesulfenamide,1-dichloro-N-[(dimethylamino)sulfonyl]-1-fluoro-N-phenyl- |
| N-{[dichloro(fluoro)methyl]sulfanyl}-N',N'-dimethyl-N-phenylsulfuric diamide |
| InChI=1/C9H11Cl2FN2O2S2/c1-13(2)18(15,16)14(17-9(10,11)12)8-6-4-3-5-7-8/h3-7H,1-2H3 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|