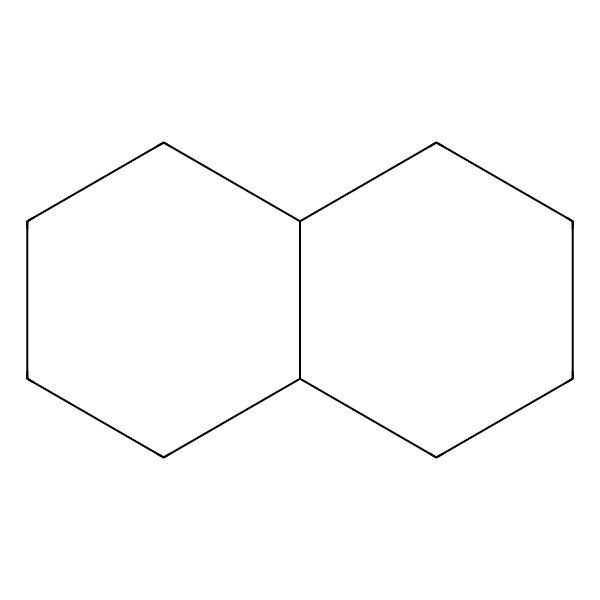
| cis-Decahydronaphthalene |
| Decalin |
| trans-Decahydronaphthalene |
| 91-17-8 |
| 493-01-6 |
| CIS-DECALIN |
| 493-02-7 |
| TRANS-DECALIN |
| Perhydronaphthalene |
| Naphthalane |
| Dekalin |
| Naphthalene, decahydro- |
| Naphthan |
| Naphthane |
| Dekalina |
| De-kalin |
| Bicyclo[4.4.0]decane |
| t-decalin |
| cis-Perhydronaphthalene |
| trans-Perhydronaphthalene |
| Naphthalene, decahydro-, cis- |
| cis-Bicyclo[4.4.0]decane |
| trans-Bicyclo[4.4.0]decane |
| Bicyclo(4.4.0)decane |
| Dekalina [Polish] |
| Decahydronaphthalene, trans- |
| c-decalin |
| Naphthalene, decahydro-, trans- |
| 1,2,3,4,4a,5,6,7,8,8a-decahydronaphthalene |
| Decahydronaphthalin |
| Dekahydronaphthalin |
| NSC 406139 |
| c-decahydronaphthalene |
| CCRIS 3410 |
| HSDB 287 |
| (4ar,8ar)-decahydronaphthalene |
| (4as,8as)-decahydronaphthalene |
| EINECS 202-046-9 |
| UN1147 |
| BRN 0878165 |
| cis-Bicyclo(4.4.0)decane |
| trans-Bicyclo(4.4.0)decane |
| UNII-88451Q4XYF |
| AI3-01256 |
| DTXSID1024912 |
| CHEBI:38853 |
| 88451Q4XYF |
| EINECS 207-770-9 |
| EINECS 207-771-4 |
| NSC 77452 |
| NSC 77453 |
| NSC-406139 |
| EC 202-046-9 |
| 3-05-00-00245 (Beilstein Handbook Reference) |
| Naphthalene-1,2,3,4,5,6,7,8-d8, decahydro-d10- |
| MFCD00004130 |
| trans decalin |
| decahydronaphtalene |
| Decalin(R) |
| Naphthalene, cis- |
| Decanhydronaphthalene |
| Naftaleno, decahidro- |
| DHN (CHRIS Code) |
| (E)-Decahydronaphthalene |
| (Z)-Decahydronaphthalene |
| Decahydronaphthalene, cis |
| DECALIN [MI] |
| Decahydronaphthalene, (E)- |
| Decahydronaphthalene, (Z)- |
| WLN: L66TJ |
| MLS001055339 |
| cis-Decahydronaphthalene, 99% |
| DTXCID304912 |
| CHEMBL1491920 |
| CHEBI:38860 |
| CHEBI:38863 |
| trans-Decahydronaphthalene, 99% |
| DTXSID00873337 |
| DTXSID90883405 |
| Decahydronaphthalene, cis + trans |
| DECAHYDRONAPHTHALENE [HSDB] |
| HMS3039O03 |
| NSC77452 |
| NSC77453 |
| STR05788 |
| Tox21_200222 |
| DECAHYDRONAPHTHALENE [WHO-DD] |
| MFCD00064189 |
| MFCD00064191 |
| NA1147 |
| NSC-77452 |
| NSC-77453 |
| NSC406139 |
| STL280340 |
| AKOS009031622 |
| AKOS016010284 |
| AKOS025295369 |
| AT32906 |
| AT32907 |
| LS-1916 |
| UN 1147 |
| CAS-91-17-8 |
| NCGC00090716-01 |
| NCGC00090716-02 |
| NCGC00257776-01 |
| BS-23504 |
| BS-23873 |
| SMR000673566 |
| Decahydronaphthalene, cis-; (cis-Decalin) |
| D0007 |
| D0008 |
| D0009 |
| D1738 |
| FT-0623872 |
| FT-0623939 |
| FT-0632224 |
| Decahydronaphthalene, trans-; (trans-Decalin) |
| A843747 |
| cis-Decahydronaphthalene, purum, >=98.0% (GC) |
| Decahydronaphthalene [UN1147] [Flammable liquid] |
| Q415454 |
| Decahydronaphthalene [UN1147] [Flammable liquid] |
| W-109334 |
| Q27118000 |
| Q27118002 |
| DECALIN (SEE ALSO TETRALIN (CAS: 119-64-2)) |
| Z104474658 |
| InChI=1/C10H18/c1-2-6-10-8-4-3-7-9(10)5-1/h9-10H,1-8H |
|
There are more than 10 synonyms. If you wish to see them all click here.
|