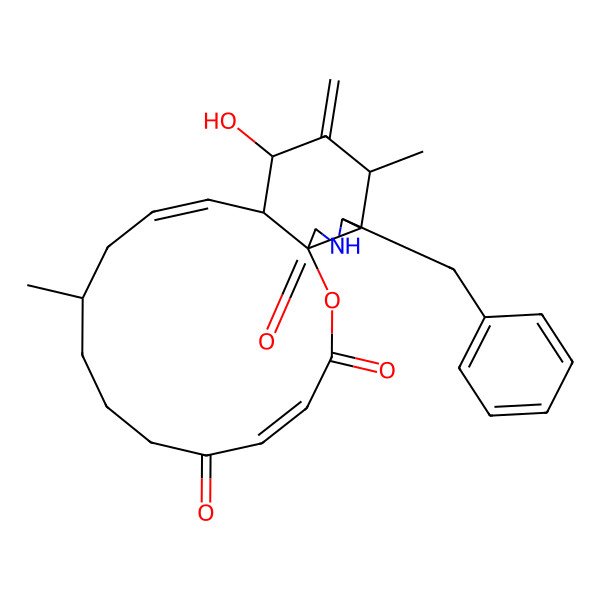
| Dehydrophomin |
| 5-Dehydrophomin |
| 5,5-Didehydrophomin |
| 14110-64-6 |
| Phomin, 5,5-didehydro- |
| BV8WQ9500E |
| EINECS 237-964-9 |
| NSC174119 |
| NSC 174119 |
| Cytochalasin A from Drechslera dematioidea |
| 24-Oxa(14)cytochalasa-6(12),13,21-triene-1,20,23-trione, 7-hydroxy-16-methyl-10-phenyl-, (7S,13E,16R,21E)- |
| 2H-Oxacyclotetradecino(2,3-d)isoindole-2,5,18-trione, 16-benzyl-6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-13-hydroxy-9,15-dimethyl-14-methylene- |
| 2H-Oxacyclotetradecino(2,3-d)isoindole-2,5,18-trione, 6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-13-hydroxy-9,15-dimethyl-14-methylene-16-(phenylmethyl)-, (9R-(3E,9R*,11E,12aS*,13S*,15S*,15aS*,16S*,18aS*))- |
| 2H-Oxacyclotetradecino(2,3-d)isoindole-2,5,18-trione, 6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-13-hydroxy-9,15-dimethyl-14-methylene-16-(phenylmethyl)-, (3E,9R,11E,12aS,13S,15S,15aS,16S,18aS)- |
| 24-Oxa[14]cytochalasa-6(12),13,21-triene-1,20,23-trione, 7-hydroxy-16-methyl-10-phenyl-, (7S,13E,16R,21E)- |
| 2H-Oxacyclotetradecino[2,3-d]isoindole-2,5,18-trione, 6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-13-hydroxy-9,15-dimethyl-14-methylene-16-(phenylmethyl)-, (3E,9R,11E,12aS,13S,15S,15aS,16S,18aS)- |
| Spectrum5_001741 |
| UNII-BV8WQ9500E |
| CHEBI:144392 |
| DTXSID401017587 |
| (1S,4E,10R,12E,14S,15S,17S,18S,19S)-19-benzyl-15-hydroxy-10,17-dimethyl-16-methylidene-2-oxa-20-azatricyclo[12.7.0.01,18]henicosa-4,12-diene-3,6,21-trione |
| 2H-Oxacyclotetradec[2,3-d]isoindole-2,5,18-trione, 16-benzyl-6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-13-hydroxy-9,15-dimethyl-14-methylene- |
| 7(S)-Hydroxy-16(R)-methyl-10-phenyl-24-oxa(14)cytochalasa-6(12),13(E),21(E)-triene-1,20,23-trione |
| 7(S)-hydroxy-16(R)-methyl-10-phenyl-24-oxa[14]cytochalasa-6(12),13(E),21(E)-triene-1,20,23-trione |
| HY-N6773 |
| CCG-39699 |
| LMPK11000001 |
| MFCD00005935 |
| AKOS030213126 |
| NCGC00388356-01 |
| benzyl-hydroxy-dimethyl-methylene-[?]trione |
| CS-0091953 |
| C19953 |
| Cytochalasin A from Helminthosporium dematioideum |
| J-007460 |
| (7S,13E,16R,21E)-7-Hydroxy-16-methyl-10-phenyl-24-oxa[14]cytochalasa-6(12),13,21-triene-1,20,23-trione |
| 16-Benzyl-13-hydroxy-9,15-dimethyl-14-methylene-8,9,10,12a,13,14,15,15a,16,17-decahydro-2H-oxacyclotetradecino[2,3-d]isoindole-2,6,18(5H,7H)-trione |
| 2H-Oxacyclotetradecino[2,3-d]isoindole-2,5,18-trione,6,7,8,9,10,12a,13,14,15,15a,16,17-dodecahydro-13-hydroxy-9,15-dimethyl-14-methylene-16-(phenylmethyl)-,(3E,9R,11E,12aS,13S,15S,15aS,16S,18aS)- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|