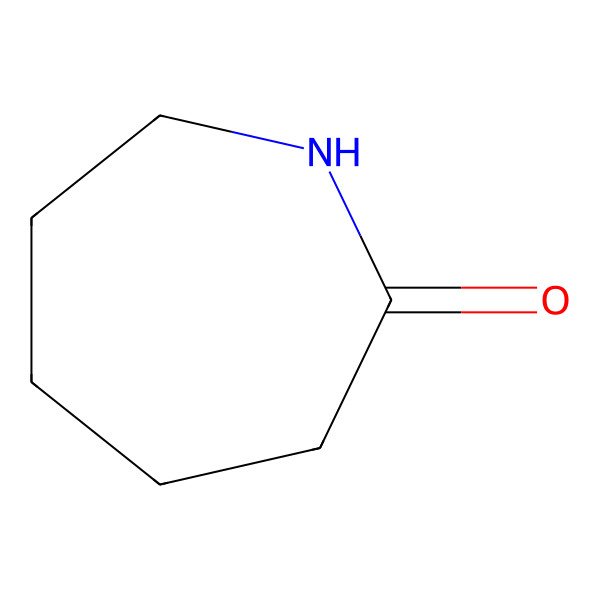
| CAPROLACTAM |
| epsilon-Caprolactam |
| 105-60-2 |
| 6-Caprolactam |
| 2-Oxohexamethylenimine |
| Aminocaproic lactam |
| 2H-Azepin-2-one, hexahydro- |
| 6-Hexanelactam |
| 2-Azacycloheptanone |
| Hexahydro-2H-azepin-2-one |
| 2-Oxohexamethyleneimine |
| Hexanolactam |
| 2-Perhydroazepinone |
| Hexahydro-2-azepinone |
| 1,6-Hexolactam |
| Hexanone isoxime |
| Caprolattame |
| 2-Ketohexamethylenimine |
| Cyclohexanone iso-oxime |
| E-Caprolactam |
| 1-Aza-2-cycloheptanone |
| Epsylon kaprolaktam |
| Hexanonisoxim |
| Kaprolaktam |
| 6-Aminocaproic acid lactam |
| Extrom 6N |
| hexannic acid |
| e-Kaprolaktam |
| Hexamethylenimine, 2-oxo- |
| Caprolactam monomer |
| 1,6-Hexalactam |
| 2-Ketohexamethyleneimine |
| Kapromine |
| Stilon |
| E-caprolactum |
| 6-Aminohexanoic acid cyclic lactam |
| Perhydroazepin-2-one |
| 2H-Azepin-7-one, hexahydro- |
| gamma-caprolactam |
| omega-caprolactum |
| Capron PK4 |
| .epsilon.-Caprolactam |
| Hexanoic acid, 6-amino-, lactam |
| NCI-C50646 |
| .omega.-Caprolactam |
| 2H-azepin-7-one,hexahydro |
| Hexanoic acid, 6-amino-, cyclic lactam |
| Aza-2-cycloheptanone |
| CCRIS 119 |
| HSDB 187 |
| EINECS 203-313-2 |
| Lactam, aminocaproic |
| NSC 117393 |
| hexanoic acid-6-amino-,lactam |
| BRN 0106934 |
| DTXSID4020240 |
| CHEBI:28579 |
| AI3-14515 |
| UNII-6879X594Z8 |
| A1030 |
| cis-Hexahydro-2-azepinone |
| NSC-117393 |
| hexahydro 2H Azepin 2 one |
| hexanoic acid-6-amino-lactam |
| DTXCID00240 |
| 6879X594Z8 |
| FEMA NO. 4235 |
| 2-Azepinone, hexahydro-, (Z)- |
| EC 203-313-2 |
| EPSILON-CAPROLACTAM-D10 |
| 5-21-06-00444 (Beilstein Handbook Reference) |
| Cyclohexanoneisooxime |
| Hexanoic acid, lactam |
| WLN: T7MVTJ |
| CAPROLACTAM (IARC) |
| CAPROLACTAM [IARC] |
| Hexanoic acid, cyclic lactam |
| CAPROLACTAM (USP-RS) |
| CAPROLACTAM [USP-RS] |
| HEXANOIC ACID,6-AMINO,LACTAM E-CAPROLACTAM |
| Caprolattame [French] |
| e-Kaprolaktam [Czech] |
| Hexanonisoxim [German] |
| Epsylon kaprolaktam [Polish] |
| CAS-105-60-2 |
| hexahydroazepin-2-one |
| ZINC ACEXAMATE IMPURITY D (EP IMPURITY) |
| ZINC ACEXAMATE IMPURITY D [EP IMPURITY] |
| MFCD00006936 |
| Caprolactam dust and vapor |
| caprolactim |
| Alkamid |
| Chemlon |
| Danamid |
| Kaprolit |
| Kaprolon |
| Metamid |
| Orgamide |
| Steelon |
| Akulon |
| Capron |
| Grilon |
| Itamid |
| Kapron |
| Stylon |
| Vidlon |
| Widlon |
| Orgamid rmnocd |
| Durethan bk |
| Ultramid bmk |
| 6-hexanolactam |
| Tarlon xb |
| Caprolon B |
| Caprolon V |
| Kaprolit B |
| Kaprolon B |
| Renyl mv |
| Tarnamid T |
| hexano-6-lactam |
| Itamide S |
| Capron B |
| Ertalon 6sa |
| Kapron A |
| Kapron B |
| Caprolactam,(S) |
| Relon P |
| epsilon caprolactam |
| Durethan bk 30S |
| Tarlon X-A |
| Tarnamid T 2 |
| Ultramid B 3 |
| Ultramid B 4 |
| Ultramid B 5 |
| 2-Azepanone # |
| Durethan bkv 30H |
| Durethan bkv 55H |
| Itamide 25 |
| Itamide 35 |
| Miramid H 2 |
| Miramid wm 55 |
| Torayca N 6 |
| epsilon -caprolactam |
| Tarnamid T 27 |
| azacycloheptan-2-one |
| Capran 77C |
| Capran 80 |
| Plaskon 8201hs |
| Plaskon xp 607 |
| Itamide 250 |
| Itamide 250G |
| Itamide 350 |
| Plaskon 201 |
| Spencer 401 |
| Spencer 601 |
| azacycloheptane-2-one |
| Sipas 60 |
| Itamid 250 |
| Maranyl F 114 |
| Maranyl F 124 |
| Maranyl F 500 |
| Nylon A1035sf |
| Plaskon 8202C |
| Capron gr 8256 |
| Capron gr 8258 |
| Plaskin 8200 |
| Plaskon 8201 |
| Plaskon 8205 |
| Plaskon 8207 |
| Plaskon 8252 |
| 6-CAPROLACTAN |
| Capron 8250 |
| Capron 8252 |
| Capron 8253 |
| Capron 8256 |
| Capron 8257 |
| Nylon cm 1031 |
| Zytel 211 |
| ?2-Oxohexamethylenimine |
| Amilan CM 1001C |
| Amilan CM 1001G |
| Amilan CM 1001 |
| Amilan CM 1011 |
| CAPROLACTAM [MI] |
| Dull 704 |
| Nylon X 1051 |
| CAPROLACTAM [HSDB] |
| ATM 2(NYLON) |
| bmse000372 |
| epsilon-Caprolactam, 99% |
| 6-Aminohexanoic acid lactam |
| Hexahydro-2H-azepine-2-one |
| SCHEMBL19610 |
| 9012-16-2 |
| CHEMBL276218 |
| PK 4 |
| HEXAMETHYLENIMINE,2-OXO- |
| NIOSH/CM3900000 |
| 1,6-HEXALACTAM [FHFI] |
| UBE 1022B |
| NSC4977 |
| 2H-AZEPIN-2-ONE,HEXAHYDRO |
| NSC-4977 |
| NSC25536 |
| STR02412 |
| Tox21_202202 |
| Tox21_300163 |
| c0432 |
| NSC-25536 |
| NSC117393 |
| AKOS000119969 |
| CM 1001 |
| CM 1011 |
| CM 1031 |
| CM 1041 |
| epsilon-Caprolactam, analytical standard |
| NCGC00247913-01 |
| NCGC00247913-02 |
| NCGC00253933-01 |
| NCGC00259751-01 |
| AM802872 |
| CM39000000 |
| FT-0623443 |
| FT-0625676 |
| EN300-19667 |
| A23500 |
| C06593 |
| D70254 |
| Q409397 |
| J-510225 |
| F0001-0110 |
| InChI=1/C6H11NO/c8-6-4-2-1-3-5-7-6/h1-5H2,(H,7,8 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|