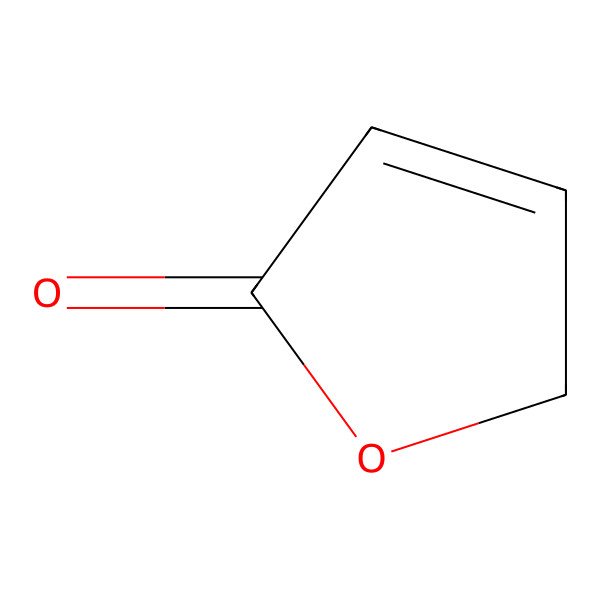
| 497-23-4 |
| Furan-2(5H)-one |
| BUTENOLIDE |
| Isocrotonolactone |
| 2-Buten-4-olide |
| 2-Butenolide |
| gamma-Crotonolactone |
| 2-Oxo-2,5-dihydrofuran |
| 2H-furan-5-one |
| Crotonolactone |
| 4-Hydroxy-2-butenoic acid lactone |
| gamma-Crotolactone |
| alpha,beta-Crotonolactone |
| 2-(5H)-furanone |
| delta,alpha,beta-Butenolide |
| gamma-Hydroxycrotonic acid lactone |
| 5H-furan-2-one |
| but-2-en-4-olide |
| 2-Butenoic acid gamma-lactone |
| 2-Butenoic acid-gamma-lactone |
| .gamma.-Crotonolactone |
| C4H4O2 |
| 2-Buten-1,4-olide |
| 2,5-dihydrofuran-2-one |
| 4-Hydroxycrotonic acid gamma-lactone |
| NSC 197009 |
| CCRIS 5722 |
| 2,5-Dihydrofuranone |
| 2-Butenoic acid, 4-hydroxy-, gamma-lactone |
| EINECS 207-839-3 |
| MFCD00005376 |
| BRN 0383585 |
| 4-Hydroxy-2-butenoic acid gamma-lactone |
| UNII-8KXK25H388 |
| CHEMBL166223 |
| CHEBI:38118 |
| HSDB 8151 |
| 2-Butenoic acid-.gamma.-lactone |
| 8KXK25H388 |
| .delta.,.alpha.,.beta.-Butenolide |
| .gamma.-Hydroxycrotonic acid lactone |
| NSC-197009 |
| 4-Hydroxy-2-butenoic acid .gamma.-lactone |
| Crotonic acid, 4-hydroxy-, .gamma.-lactone |
| 2-Butenoic acid, 4-hydroxy-, .gamma.-lactone |
| 5-17-09-00112 (Beilstein Handbook Reference) |
| Crotonic acid, 4-hydroxy-, gamma-lactone (6CI,7CI) |
| .DELTA..alpha.,.beta.-Butolide |
| 2-Butenoic acid, .gamma.-lactone |
| .DELTA..alpha.,.beta.-Butenolide |
| g-Crotonolactone |
| ?-Crotonolactone |
| 5H-furanone |
| 2-B4O |
| 2_5H_furanone |
| CRATONE |
| .gamma.-Crotolactone |
| 5 H-Furan-2-one |
| (5H)-furan-2-one |
| alpha,alpha,beta-Butolide |
| 2(5H)FURANONE |
| .BETA.-CROTONOLACTONE |
| 2(5H)-Furanone, 98% |
| laquo gammaRaquo -crotolactone |
| 4-Hydroxycrotonic acid lactone |
| .alpha.,.beta.-Crotonolactone |
| laquo gammaRaquo -crotonolactone |
| DTXSID7075422 |
| FEMA NO. 4138 |
| VIHAEDVKXSOUAT-UHFFFAOYSA- |
| BCP15247 |
| NSC51296 |
| BBL103594 |
| BDBM50360798 |
| NSC-51296 |
| NSC197009 |
| STL557404 |
| AKOS000268618 |
| CS-W008270 |
| HY-W008270 |
| laquo deltaRaquo ,alpha,beta-butenolide |
| SB60895 |
| Crotonic acid, 4-hydroxy-, gamma-lactone |
| LS-70308 |
| SY017893 |
| 2-Butenoic acid-laquo gammaRaquo -lactone |
| 2-Oxo-2,5-dihydrofuran(2-[5H]-furanone) |
| 5-OXO-2,5-DIHYDROFURAN-3-YL ESTER |
| A7404 |
| AM20100212 |
| FT-0608698 |
| EN300-80159 |
| laquo gammaRaquo -hydroxycrotonic acid lactone |
| C17601 |
| 2(5H)-Furanone (laquo gammaRaquo -crotonolactone) |
| 4-Hydroxy-2-butenoic acid laquo gammaRaquo -lactone |
| J-506134 |
| Q4596886 |
| Crotonic acid, 4-hydroxy-, laquo gammaRaquo -lactone |
| 2-Butenoic acid, 4-hydroxy-, laquo gammaRaquo -lactone |
| Z1219048777 |
| 4-HYDROXY-2-BUTENOIC ACID .GAMMA.-LACTONE [FHFI] |
| InChI=1/C4H4O2/c5-4-2-1-3-6-4/h1-2H,3H2 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|