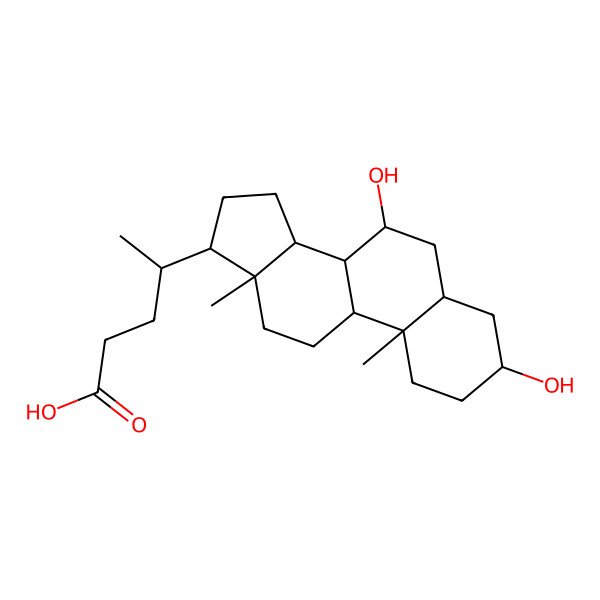
| ursodiol |
| 128-13-2 |
| Actigall |
| ursodeoxycholate |
| UDCA |
| Ursofalk |
| Ursolvan |
| UrSO |
| Delursan |
| Ursodesoxycholic acid |
| Urso Forte |
| Destolit |
| Ursochol |
| Cholit-ursan |
| Litursol |
| Solutrat |
| Ursobilin |
| Ursodamor |
| Arsacol |
| Deursil |
| Lyeton |
| Ursacol |
| Peptarom |
| Urso DS |
| UDCS |
| 3alpha,7beta-Dihydroxy-5beta-cholan-24-oic acid |
| Ursosan |
| Ursacholic Acid |
| Urso 250 |
| Ursodiol [USAN] |
| Deoxyursocholic Acid |
| Antigall |
| Urosiol |
| 3-alpha,7-beta-Dioxycholanic acid |
| 7-beta-Hydroxylithocholic acid |
| Ursocholic acid, deoxy- |
| Ursodeoxycholicacid |
| 3-alpha,7-beta-Dihydroxycholanic acid |
| Ursodexycholic Acid |
| Acido ursodeossicolico [Italian] |
| Acido ursodeoxicolico |
| Acido ursodeossicolico |
| Acido ursodeoxicolico [INN-Spanish] |
| Acidum ursodeoxycholicum [INN-Latin] |
| Acide ursodesoxycholique |
| Acide ursodesoxycholique [INN-French] |
| Acidum ursodeoxycholicum |
| Ursonorm |
| NSC 683769 |
| 3-alpha,7-beta-Dihydroxy-5-beta-cholanoic acid |
| Sodium Ursodeoxycholate |
| (3alpha,5beta,7beta)-3,7-dihydroxycholan-24-oic acid |
| Actigall (TN) |
| Ursodiol (USP) |
| NSC 657950 |
| BRN 3219888 |
| 3,7-Dihydroxycholan-24-oic acid |
| CCRIS 5502 |
| (4R)-4-[(3R,5S,7S,8R,9S,10S,13R,14S,17R)-3,7-dihydroxy-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid |
| Urso (TN) |
| Ursodiol [USAN:USP] |
| EINECS 204-879-3 |
| Ursodeoxycholic acid [INN] |
| ursocol |
| CHEMBL1551 |
| UNII-724L30Y2QR |
| MLS000028461 |
| CHEBI:9907 |
| 17-beta-(1-Methyl-3-carboxypropyl)etiocholane-3-alpha,7-beta-diol |
| 724L30Y2QR |
| 3 alpha,7 beta-Dihydroxy-5 beta-cholan-24-oic Acid |
| NSC-683769 |
| 5beta-Cholanic Acid-3alpha,7beta-diol |
| SMR000058403 |
| Cholan-24-oic acid, 3,7-dihydroxy-, (3alpha,5beta,7beta)- |
| 3alpha,7beta-Dihydroxy-5beta-cholanic acid |
| U0030 |
| Cholan-24-oic acid, 3,7-dihydroxy-, (3.alpha.,5.beta.,7.beta.)- |
| EC 204-879-3 |
| U-9000 |
| (R)-4-((3R,5S,7S,8R,9S,10S,13R,14S,17R)-3,7-dihydroxy-10,13-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentanoic acid |
| 4-10-00-01604 (Beilstein Handbook Reference) |
| IU5 |
| 5-beta-Cholan-24-oic acid, 3-alpha,7-beta-dihydroxy- |
| (3alpha,5beta,7beta,8xi)-3,7-dihydroxycholan-24-oic acid |
| Cholan-24-oic acid, 3,7-dihydroxy-, (3-alpha,5-beta,7-beta)- |
| Dom-ursodiol c |
| PHL-ursodiol c |
| PMS-ursodiol c |
| 7beta-Hydroxylithocholic acid |
| SR-01000737091 |
| MFCD00003680 |
| Ursodexycholate |
| Paptarom |
| Udiliv |
| Desol |
| Urdes |
| Urosdesoxycholate |
| 5beta-Cholan-24-oic acid-3alpha,7beta-diol |
| Ursodesoxycholicacid |
| Ursodeoxycholoc acid |
| Deursil (TN) |
| Ursosan (TN) |
| Urosdesoxycholic acid |
| Urso Forte (TN) |
| URSODIOL [INCI] |
| URSODIOL [MI] |
| Ursodiol (Actigal Urso) |
| URSODIOL [VANDF] |
| Prestwick0_000958 |
| Prestwick1_000958 |
| Prestwick2_000958 |
| Prestwick3_000958 |
| 7A-Hydroxylithocholic acid |
| D0G3SH |
| URSODIOL [USP-RS] |
| (3-alpha,5-beta,7-beta)-3,7-Dihydroxycholan-24-oic acid |
| SCHEMBL27200 |
| BSPBio_000956 |
| 7bet.-Hydroxylithocholic acid |
| cid_31401 |
| MLS001066373 |
| MLS002548885 |
| SPBio_003105 |
| 17-beta-(1-methyl-3-carboxypropyl)etiocholane-3-alpha,7beta-diol |
| Ursodeoxycholic acid, >=99% |
| URSODIOL [ORANGE BOOK] |
| BPBio1_001052 |
| GTPL7104 |
| 7.beta.-Hydroxylithocholic acid |
| Ursodiol (Ursodeoxycholic Acid) |
| DTXSID6023731 |
| URSODIOL [USP MONOGRAPH] |
| BDBM53721 |
| HMS1570P18 |
| HMS2097P18 |
| HMS2233L14 |
| HMS3259A13 |
| HMS3714P18 |
| Ursodeoxycholic acid (JP15/INN) |
| Ursodeoxycholic acid (JP17/INN) |
| URSODEOXYCHOLIC ACID [JAN] |
| 3alpha,7beta-dihydroxycholanic acid |
| 3a,7b-dihydroxy-5b-cholan-24-oate |
| HB4645 |
| LMST04010033 |
| s1643 |
| URSODEOXYCHOLIC ACID [MART.] |
| URSODEOXYCHOLIC ACID [WHO-DD] |
| 5A-Cholan-24-oic acid-3A,7A-diol |
| AKOS015955898 |
| CCG-220958 |
| CS-1932 |
| CS-O-30812 |
| DB01586 |
| KS-5243 |
| NC00487 |
| SMP2_000012 |
| 3.alpha.,7.beta.-Dihydroxycholanic acid |
| 3A,7A-Dihydroxy-5A-holan-24-oic acid |
| 3a,7b-dihydroxy-5b-cholan-24-oic acid |
| NCGC00179363-01 |
| NCGC00179363-12 |
| AC-18919 |
| CAS#128-13-2 |
| CPD000058403 |
| HY-13771 |
| LS-53033 |
| NCI60_028904 |
| URSODEOXYCHOLIC ACID [EP IMPURITY] |
| URSODEOXYCHOLIC ACID [EP MONOGRAPH] |
| AB00513977 |
| (3a,5b,7b)-3,7-dihydroxy-cholan-24-oate |
| 3alpha, 7beta-dihydroxy-5beta-cholanoic acid |
| 5bet.-Cholan-24-oic acid-3alp.,7bet.-diol |
| (3a,5b,7b)-3,7-dihydroxycholan-24-oic acid |
| C07880 |
| D00734 |
| EN300-373707 |
| ((3a,5b,7b)-3,7-Dihydroxycholan-24-oic acid |
| (3a,5b,7b)-3,7-dihydroxy-cholan-24-oic acid |
| AB00513977-09 |
| AB00513977_10 |
| 3alp.,7bet.-Dihydroxy-5bet.-cholan-24-oic acid |
| A905413 |
| Q241374 |
| 3.alpha.,7.beta.-Dihydroxy-5.beta.-cholanic acid |
| J-005566 |
| J-650210 |
| SR-01000737091-3 |
| SR-01000737091-4 |
| BRD-K15697815-001-16-2 |
| 3.alpha.,7.beta.-Dihydroxy-5.beta.-cholan-24-oic acid |
| Z2588039022 |
| 3.ALPHA.,7.BETA.-DIHYDROXY-5B-CHOLAN-24-OIC ACID |
| 5.beta.-Cholan-24-oic acid, 3.alpha.,7.beta.-dihydroxy- |
| Ursodiol, United States Pharmacopeia (USP) Reference Standard |
| Ursodeoxycholic acid, British Pharmacopoeia (BP) Reference Standard |
| Ursodeoxycholic acid, European Pharmacopoeia (EP) Reference Standard |
| 17.beta.-(1-Methyl-3-carboxypropyl)etiocholane-3.alpha.,7.beta.-diol |
| 3,7-Dihydroxycholan-24-oic acid-, (3.alpha.,5.beta.,7.beta.)- # |
| Cholan-24-oic acid, 3,7-dihydroxy-, (3-alpha,5-beta,7-beta)-(9CI) |
| Ursodeoxycholic acid, 500 mug/mL in methanol, certified reference material |
| Ursodiol, Pharmaceutical Secondary Standard; Certified Reference Material |
| Ursodeoxycholic acid for system suitability, European Pharmacopoeia (EP) Reference Standard |
| Ursodeoxycholic acid, UDCA, Ursosan, Ursofalk, Urso Forte, Udiliv, Ursodiol |
| (4R)-4-[(1R,3aS,3bR,4S,5aS,7R,9aS,9bS,11aR)-4,7-dihydroxy-9a,11a-dimethyl-hexadecahydro-1H-cyclopenta[a]phenanthren-1-yl]pentanoic acid |
| (4R)-4-[(1S,2S,5R,7S,9S,10R,11S,14R,15R)-5,9-Dihydroxy-2,15-dimethyltetracyclo[8.7.0.02,1.011,1]heptadecan-14-yl]pentanoic acid |
| (4R)-4-[(3R,5S,7S,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3,7-bis(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid |
| (4R)-4-[(3R,5S,7S,8R,9S,10S,13R,14S,17R)-3,7-dihydroxy-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoicacid |
| (4R)-4-[(3R,5S,7S,8R,9S,10S,13R,14S,17R)-3,7-dihydroxy-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]valeric acid |
| 108609-27-4 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|