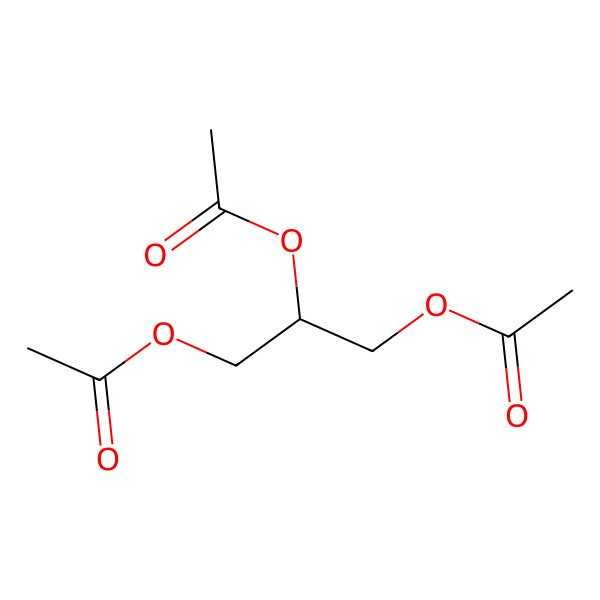
| 102-76-1 |
| Glyceryl triacetate |
| Glycerol triacetate |
| Enzactin |
| Glycerin triacetate |
| Triacetine |
| Triacetylglycerol |
| Fungacetin |
| Glyped |
| Vanay |
| Triacetyl glycerine |
| Kesscoflex TRA |
| Kodaflex triacetin |
| 1,2,3-Propanetriol, triacetate |
| 1,2,3-triacetoxypropane |
| Acetin, tri- |
| Triacetina |
| Triacetinum |
| 1,2,3-Propanetriol triacetate |
| propane-1,2,3-triyl triacetate |
| Ujostabil |
| 1,2,3-Propanetriol, 1,2,3-triacetate |
| Triacetyl glycerin |
| Triacetyl glycerol |
| Estol 1581 |
| Triacetin [INN] |
| FEMA No. 2007 |
| 1,2,3-Propanetriyl triacetate |
| 1,2,3-Triacetylglycerol |
| Glyceryltriacetate |
| FEMA Number 2007 |
| NSC 4796 |
| 2,3-diacetyloxypropyl acetate |
| HSDB 585 |
| triacetylglycerin |
| Triacetine [INN-French] |
| Triacetinum [INN-Latin] |
| Ins no.1518 |
| Triacetina [INN-Spanish] |
| Acetic, 1,2,3-propanetriyl ester |
| NSC-4796 |
| EINECS 203-051-9 |
| UNII-XHX3C3X673 |
| Glycerine triacetate |
| Ins-1518 |
| BRN 1792353 |
| CCRIS 9355 |
| CHEBI:9661 |
| XHX3C3X673 |
| DTXSID3026691 |
| AI3-00661 |
| C9H14O6 |
| Triacetin (USP/INN) |
| E1518 |
| ENZACTIN (TN) |
| E-1518 |
| 1,2,3-triacetyl-glycerol |
| TRIACETIN (GLYCEROL TRIACETATE) |
| 2-(Acetyloxy)-1-[(acetyloxy)methyl]ethyl acetate |
| Triacetin [USP:INN:BAN] |
| 1,2,3-triacetyl-sn-glycerol |
| DTXCID906691 |
| EC 203-051-9 |
| 4-02-00-00253 (Beilstein Handbook Reference) |
| NCGC00091612-04 |
| TRIACETIN (II) |
| TRIACETIN [II] |
| Triacetin (1,2,3-Propanetriol triacetate) |
| TRIACETIN (MART.) |
| TRIACETIN [MART.] |
| E 1518 |
| TRIACETIN (USP-RS) |
| TRIACETIN [USP-RS] |
| TRIACETIN (EP MONOGRAPH) |
| TRIACETIN [EP MONOGRAPH] |
| TRIACETIN (USP MONOGRAPH) |
| TRIACETIN [USP MONOGRAPH] |
| Triacetyl-glycerol |
| CAS-102-76-1 |
| 2-(Acetyloxy)-1-((acetyloxy)methyl)ethyl acetate |
| Enzacetin |
| Euzactin |
| Fungacet |
| Motisil |
| Blekin |
| tri-acetin |
| Acetin-tri |
| Triacetin, CP |
| Edenor GTA |
| Triacetin, FCC |
| Triacetin, USP |
| (tri-)Acetin |
| 3-Triacetoxypropane |
| MFCD00008716 |
| Priacetin 1580 |
| Priacetin 1581 |
| Triacetin, 99% |
| Triacetinum (Latin) |
| triacetato de glicerina |
| 123-Triacetoxypropane |
| Spectrum_000881 |
| TRIACETIN [FCC] |
| TRIACETIN [MI] |
| TRIACETIN [FHFI] |
| TRIACETIN [HSDB] |
| TRIACETIN [INCI] |
| Acetin tri-(6CI8CI) |
| Spectrum2_000939 |
| Spectrum3_001368 |
| Spectrum4_000362 |
| Spectrum5_001376 |
| TRIACETIN [VANDF] |
| D0Q6DX |
| Triacetin, >=99.5% |
| SCHEMBL3870 |
| TRIACETIN [WHO-DD] |
| BSPBio_002896 |
| Glycerol triacetate tributyrin |
| KBioGR_000823 |
| KBioSS_001361 |
| MLS002152946 |
| 1,3-Propanetriol, triacetate |
| DivK1c_000740 |
| Glyceryl triacetate, >=99% |
| SPECTRUM1500585 |
| Triacetin, analytical standard |
| SPBio_000878 |
| 1,2,3 Propanetriol triacetate |
| Triacetin, 99%, FCC, FG |
| 1,2,3-propanediol triethanoate |
| CHEMBL1489254 |
| Triacetin; (Triacetyl glycerin) |
| 1,2,3-Propanotriol, triacetato |
| FEMA 2007 |
| HMS502E22 |
| KBio1_000740 |
| KBio2_001361 |
| KBio2_003929 |
| KBio2_006497 |
| KBio3_002116 |
| DRA 150 |
| NSC4796 |
| NINDS_000740 |
| HMS1921G05 |
| HMS2092O09 |
| HMS2232I22 |
| Pharmakon1600-01500585 |
| Triacetin, >=99%, natural, FG |
| HY-B0896 |
| Tox21_111155 |
| Tox21_201745 |
| Tox21_300111 |
| WLN: 1VO1YOV1 & 1OV1 |
| 123-Propanetriol triacetate (9CI) |
| CCG-39680 |
| LMGL03012615 |
| NSC757364 |
| s4581 |
| Triacetin, 8CI, BAN, INN, USAN |
| 1,2,3-Propanetriol triacetate, 9CI |
| AKOS009028851 |
| Tox21_111155_1 |
| Glyceryl triacetate, >=99.0% (GC) |
| LS-2356 |
| NSC-757364 |
| 1,2,3-Propanetriol 1,2,3-triacetate |
| 1,3-bis(acetyloxy)propan-2-yl acetate |
| IDI1_000740 |
| Acetin, tri-, 1,2,3-Triacetoxipropano |
| NCGC00091612-01 |
| NCGC00091612-02 |
| NCGC00091612-03 |
| NCGC00091612-05 |
| NCGC00091612-06 |
| NCGC00091612-07 |
| NCGC00091612-09 |
| NCGC00254207-01 |
| NCGC00259294-01 |
| 1,2,3-Propanotriol, 1,2,3-triacetato |
| SMR001224538 |
| SBI-0051540.P002 |
| FT-0626753 |
| G0086 |
| EN300-19216 |
| D00384 |
| E75962 |
| Q83253 |
| AB00052112_06 |
| A800614 |
| SR-05000002079 |
| J-000781 |
| SR-05000002079-1 |
| 2-(Acetyloxy)-1-[(acetyloxy)methyl]ethyl acetate # |
| Z104473192 |
| Triacetin, GTA F.G (1,2,3-PROPANETRIOL TRIACETATE) |
| Triacetin, United States Pharmacopeia (USP) Reference Standard |
| Triacetin, Pharmaceutical Secondary Standard; Certified Reference Material |
| 1,2,3-Propanetriol triacetate; Glycerol Triacetate, USP Grade(1.03000); TRIACETINE; Glycerol triacetate; Glyceryl triacetate; propane-1,2,3-triyl triacetate |
| InChI=1/C9H14O6/c1-6(10)13-4-9(15-8(3)12)5-14-7(2)11/h9H,4-5H2,1-3H |
|
There are more than 10 synonyms. If you wish to see them all click here.
|