| 17673-25-5 |
| UNII-XUZ76S9127 |
| CHEBI:8116 |
| 4alpha-Phorbol |
| 4beta-Phorbol |
| 4-beta-Phorbol |
| XUZ76S9127 |
| 4,9,12beta,13,20-pentahydroxy-1,6-tigliadien-3-on |
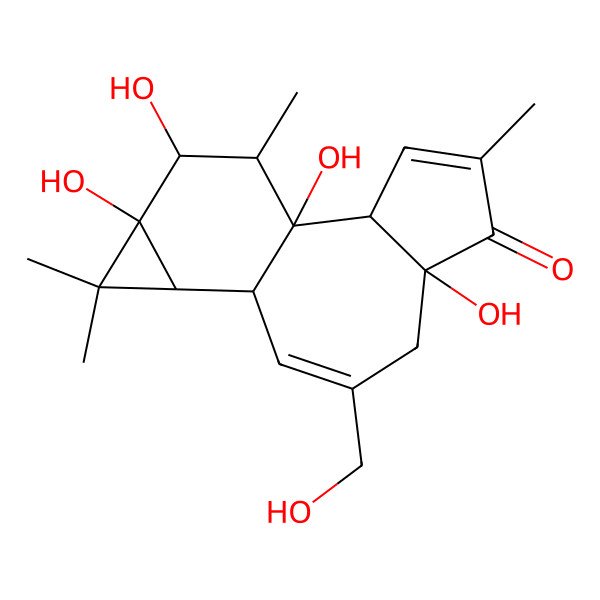
| (1aR,1bS,4aR,7aS,7bS,8R,9R,9aS)-4a,7b,9,9a-tetrahydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-1,1a,1b,4,4a,7a,7b,8,9,9a-decahydro-5H-cyclopropa[3,4]benzo[1,2-e]azulen-5-one |
| 4,9,12-beta,13,20-Pentahydroxy-1,6-tigliadien-3-on |
| 4,9,12beta,13,20-pentahydroxytiglia-1,6-dien-3-one |
| 5H-Cyclopropa[3,4]benz[1,2-e]azulen-5-one,1,1a,1b,4,4a,7a,7b,8,9,9a-decahydro-4a,7b,9,9a-tetrahydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-,(1aR,1bS,4aR,7aS,7bS,8R,9R,9aS)- |
| CCRIS 694 |
| HSDB 4328 |
| 4-Phorbol |
| 4-.beta.-Phorbol |
| NSC 154778 |
| (1aR,1bS,4aR,7aS,7bS,8R,9R,9aS)-4a,7b,9,9a-Tetrahydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-1b,4,4a,7a,7b,8,9,9a-octahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-5(1aH)-one |
| 5H-Cyclopropa(3,4)benz(1,2-e)azulen-5-one, 1,1a-beta,1b-alpha,4,4a,7a-beta,7b,8,9,9a-decahydro-4a-alpha,7b-alpha,9-beta,9a-alpha-tetrahydroxy-3-(hydroxymethyl)-1,1,6,8-alpha-tetramethyl- |
| 4b-phorbol |
| NSC-154778 |
| 1,1a,1b,4,4a,7a,7b,8,9,9a-Decahydro-4a,7b,9,9a-tetrahydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5H-cyclopropa(3,4)benz(1,2-e)azulen-5-one (1aR-(1aalpha,1bbeta,4abeta,7aalpha,7balpha,8alpha,9beta,9aalpha))- |
| 5H-Cyclopropa(3,4)benz(1,2-e)azulen-5-one, 1,1a,1b,4,4a,7a,7b,8,9,9a-decahydro-4a,7b,9,9a-tetrahydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-, (1aR,1bS,4aR,7aS,7bS,8R,9R,9aS)- |
| 5H-Cyclopropa(3,4)benz(1,2-e)azulen-5-one, 1,1a,1b,4,4a,7a,7b,8,9,9a-decahydro-4a,7b,9,9a-tetrahydroxy-3-(hydroxymethyl)-1,6,8-tetramethyl-, (1aR-(1aalpha,1beta,4abeta,7aalpha,7balpha,8alpha,9beta,9aalpha))- |
| 5H-Cyclopropa(3,4)benz(1,2-e)azulen-5-one, 1,1aalpha,1bbeta,4,4a,7aalpha,7b,8,9,9a-decahydro-4abeta,7balpha,9beta,9a alpha-tetrahydroxy-3-(hydroxymethyl)-1,1,6,8alpha-tetramethyl-, (+)- |
| 5H-Cyclopropa[3,4]benz[1,2-e]azulen-5-one, 1,1a,1b,4,4a,7a,7b,8,9,9a-decahydro-4a,7b,9,9a-tetrahydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-, (1aR,1bS,4aR,7aS,7bS,8R,9R,9aS)- |
| (+)-phorbol |
| 4.b.-Phorbol |
| tetrahydroxy-(hydroxymethyl)-tetramethyl-[?]one |
| PHORBOL [HSDB] |
| PHORBOL [MI] |
| SCHEMBL14937 |
| CHEMBL124518 |
| DTXSID2021155 |
| QGVLYPPODPLXMB-UBTYZVCOSA-N |
| 26241-63-4 |
| 5H-Cyclopropa[3,4]benz[1,2-e]azulen-5-one, 1,1a.alpha.,1b.beta.,4,4a,7a.alpha.,7b,8,9,9a-decahydro-4a.beta.,7b.alpha.,9.beta.,9a.alpha.-tetrahydroxy-3-(hydroxymethyl)-1,1,6,8.alpha.-tetramethyl-, ()- |
| 5H-Cyclopropa[3,4]benz[1,2-e]azulen-5-one, 1,1a.beta.,1b.alpha.,4,4a,7a.beta.,7b,8,9,9a-decahydro-4a.alpha.,7b.alpha.,9.beta.,9a.alpha.-tetrahydroxy-3-(hydroxymethyl)-1,1,ster |
| EX-A3092 |
| HY-N2147 |
| MFCD00065450 |
| AKOS024418764 |
| CCG-268216 |
| CS-6052 |
| LMPR0104330001 |
| SMP2_000151 |
| AC-33956 |
| BS-16283 |
| P2732 |
| S9389 |
| C09155 |
| A881502 |
| SR-05000002211 |
| J-011221 |
| Q2705359 |
| SR-05000002211-2 |
| 4beta,9alpha,12beta,13alpha,20-Pentahydroxytiglia-1,6-dien-3-one |
| 5-[2-(DIETHYLAMINO)ETHYL]-3-PHENYL-1,2,4-OXADIAZOLECITRATESALT |
| (1AR,1BS,4AR,7AS,7BS,8R,9R,9AS)-1,1A,1B,4,4A,7A,7B,8,9,9A-DECAHYDRO-4A,7B,9,9A-TETRAHYDROXY-3-(HYDROXYMETHYL)-1,1,6,8-TETRAMETHYL-5H-CYCLOPROPA(3,4)BENZ(1,2-E)AZULEN-5-ONE |
| (1S,2S,6R,10S,11R,13S,14R,15R)-1,6,13,14-tetrahydroxy-8-(hydroxymethyl)-4,12,12,15-tetramethyltetracyclo[8.5.0.02,6.011,13]pentadeca-3,8-dien-5-one |
|
There are more than 10 synonyms. If you wish to see them all click here.
|