| 99-82-1 |
| 6069-98-3 |
| cis-1-isopropyl-4-methylcyclohexane |
| 1678-82-6 |
| trans-1-isopropyl-4-methylcyclohexane |
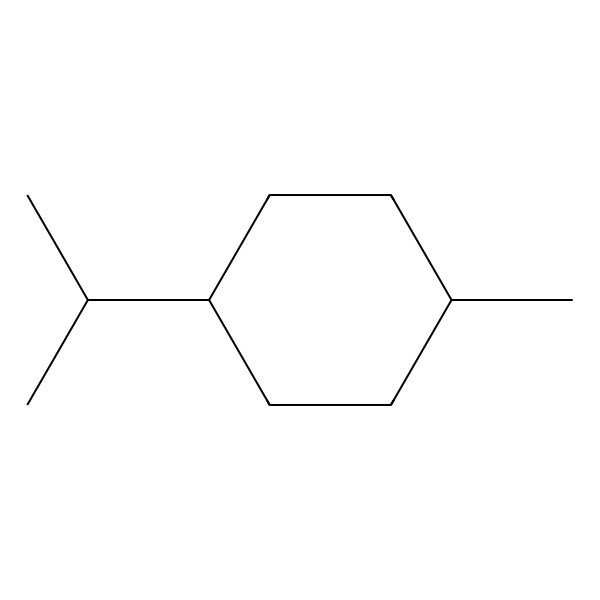
| 1-ISOPROPYL-4-METHYLCYCLOHEXANE |
| cis-p-Menthane |
| trans-p-Menthane |
| p-Menthane, trans- |
| p-Menthane, cis- |
| 1-iso-Propyl-4-methylcyclohexane |
| Cyclohexane, 1-methyl-4-(1-methylethyl)-, trans- |
| p-Menthan |
| Cyclohexane, 1-methyl-4-(1-methylethyl)-, cis- |
| 1-Methyl-trans-4-isopropylcyclohexane |
| Cyclohexane, 1-methyl-4-(1-methylethyl)- |
| 1-methyl-4-propan-2-ylcyclohexane |
| para-menthane |
| 1-Isopropyl-cis-4-methylcyclohexane |
| 1-Methyl-cis-4-isopropylcyclohexane |
| CCRIS 4664 |
| 1-Methyl-4-(1-methylethyl)-cyclohexane |
| HSDB 5328 |
| CGW5GN8TXU |
| EINECS 202-790-4 |
| NSC 73978 |
| NSC 73979 |
| 1-methyl-4-(propan-2-yl)cyclohexane |
| AI3-24486 |
| 1-methyl-4-isopropylcyclohexane |
| p-Menthane, Z- |
| trans-1-Methyl-4-isopropylcyclohexane |
| menthane |
| (1r,4r)-1-Isopropyl-4-methylcyclohexane |
| EC 202-790-4 |
| cis-1-Methyl-4-isopropylcyclohexane |
| p-Menthane E |
| 1-Isopropyl-4-methyl-cyclohexane, cis |
| PARA-MENTHANE (2,2-D2) |
| CIS-PARA-MENTHANE(3,3-D2) |
| 1-Methyl-4-(1-methylethyl)-cyclohexane, cis |
| cis-Hexahydro-p-cymene |
| trans-menthane |
| cis-menthane |
| Cyclohexane, cis- |
| UNII-CGW5GN8TXU |
| WOODY RIVER 10 |
| HU0VZO1K2G |
| 1-isopropyl-4-methylcyclohexan |
| DTXSID9025530 |
| CHEBI:25826 |
| DTXSID30884219 |
| DTXSID50883709 |
| CFJYNSNXFXLKNS-AOOOYVTPSA-N |
| CFJYNSNXFXLKNS-MGCOHNPYSA-N |
| NSC73978 |
| NSC73979 |
| MFCD00043477 |
| MFCD00070477 |
| NSC-73978 |
| NSC-73979 |
| Cis-1-isopropyl-4-methyl cyclohexane |
| AKOS006229182 |
| AKOS015838428 |
| AKOS015838431 |
| 1-methyl-4-(1-methylethyl)cyclohexane |
| LS-2070 |
| Trans-1-isopropyl-4-methyl cyclohexane |
| 1-Isopropyl-4-methyl-cyclohexane, trans |
| Cyclohexane, 1-methyl-4-isopropyl, trans |
| LS-56953 |
| FT-0600340 |
| FT-0695075 |
| FT-0695076 |
| I0284 |
| 1-ISOPROPYL-4-METHYLCYCLOHEXANE [HSDB] |
| 1-Methyl-4-(1-methylethyl)-cyclohexane, trans |
| 4-METHYL-1-(1-METHYLETHYL)CYCLOHEXANE |
| (1r,4r)-1-methyl-4-(propan-2-yl)cyclohexane |
| (1s,4s)-1-methyl-4-(propan-2-yl)cyclohexane |
| (1S,4S)-1-ISOPROPYL-4-METHYLCYCLOHEXANE |
| J-010403 |
| Q2043490 |
| W-100017 |
| InChI=1/C10H20/c1-8(2)10-6-4-9(3)5-7-10/h8-10H,4-7H2,1-3H |
|
There are more than 10 synonyms. If you wish to see them all click here.
|