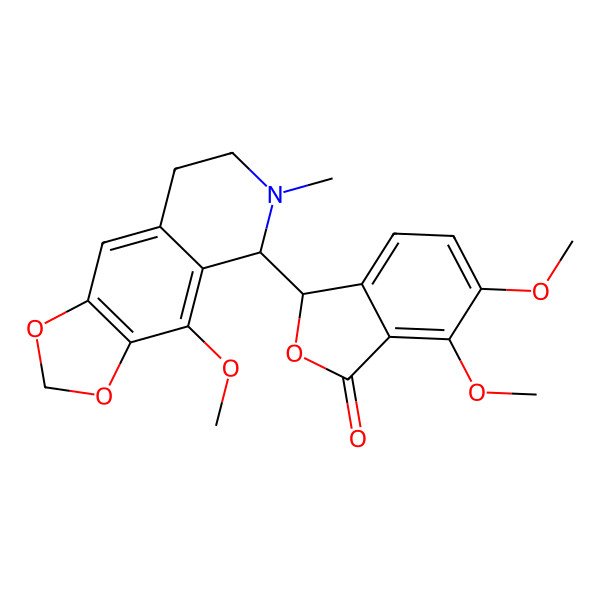
| 128-62-1 |
| NARCOTINE |
| Tusscapine |
| Methoxyhydrastine |
| Terbenol |
| Capval |
| Coscopin |
| Narcompren |
| Narcosine |
| Vadebex |
| (-)-Narcotine |
| Longatin |
| Narcotin |
| Narkotin |
| Noscapalin |
| Noscapin |
| Opianine |
| Opianin |
| Narcotussin |
| Nectadon |
| Nicolane |
| Nipaxon |
| Lyobex |
| Opian |
| alpha-Narcotine |
| O-Methylnarcotoline |
| Coscotabs |
| Noscapinum |
| Key-tusscapine |
| Longactin |
| Noscapal |
| Noscapina |
| Hederix (free base) |
| (-)-alpha-narcotine |
| L-alpha-Narcotine |
| Narcotinum |
| Noscopine |
| Noskapin |
| (-)-.alpha.-Narcotine |
| NSC 5366 |
| NSC-5366 |
| L-alpha-Noscapine |
| 8-Methoxyhydrastin |
| .alpha.-Narcotine |
| Coscopin (VAN) |
| dl-Narcotine |
| (+-)-Noscapine |
| (+/-)-Noscapine |
| L-.alpha.-Narcotine |
| (-)-alpha-Norcotine |
| (+-)-alpha-Narcotine |
| Noscapinum [INN-Latin] |
| (S,R)-Noscapine |
| Noscapina [INN-Spanish] |
| UNII-A4C6WE7BZN |
| A4C6WE7BZN |
| UNII-8V32U4AOQU |
| 8V32U4AOQU |
| alpha-Gnoscopine |
| CCRIS 9304 |
| (S)-6,7-dimethoxy-3-((R)-4-methoxy-6-methyl-5,6,7,8-tetrahydro-[1,3]dioxolo[4,5-g]isoquinolin-5-yl)isobenzofuran-1(3H)-one |
| DTXSID4023385 |
| CHEBI:73237 |
| HSDB 3372 |
| Noscapine [USP:INN:BAN:JAN] |
| Noscapine dl-form |
| NSC5366 |
| EINECS 204-899-2 |
| (3S)-6,7-dimethoxy-3-[(5R)-4-methoxy-6-methyl-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-2-benzofuran-1(3H)-one |
| (3S)-6,7-dimethoxy-3-[(5R)-4-methoxy-6-methyl-7,8-dihydro-5H-[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-3H-2-benzofuran-1-one |
| Noscapine HCl |
| BRN 0099933 |
| (3S)-6,7-dimethoxy-3-[(5R)-4-methoxy-6-methyl-2H,5H,6H,7H,8H-[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-1,3-dihydro-2-benzofuran-1-one |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-((5R)-5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo(4,5-g)isoquinolin-5-yl)-, (3S)-rel- |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo(4,5-g)-isoquinolin-5-yl), (S-(R*,S*))- |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo(4,5-g)isoquinolin-5-yl)-, (R*,S*)- |
| 4-27-00-06838 (Beilstein Handbook Reference) |
| DTXCID303385 |
| 6035-40-1 |
| C22H23NO7 |
| Narcotine alkaloid |
| .alpha.-Gnoscopine |
| NSC 96350 |
| Noscapine (narcotine) |
| (-)-3-(2-Methyl-6,7-methylendioxy-8-methoxy-1-isochinolyl)-6,7-dimethoxyphthalid |
| Narcotine, (+-)- |
| 5-(6,7-Dimethoxyphthalidyl)-5,6,7,8-tetrahydro-4-methoxy-8-methyl-1,3-dioxolo(4,5-g)isoquinoline |
| L-alpha-2-Methyl-8-methoxy-6,7-methylenedioxy-1-(6,7-dimethoxy-3-phthalidyl)-1,2,3,4-tetrahydroisoquinaline |
| SMR000059119 |
| Narcotine, (.+.)- |
| (-)-.alpha.-Norcotine |
| Noscapine [INN:BAN:JAN] |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo(4,5-g)isoquinolin-5-yl)-, (S-(R*,S*))- |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo[4,5-g]isoquinolin-5-yl)-, [S-(R*,S*)]- |
| C22-H23-N-O7 |
| NSC-96350 |
| methoxyhydra-stine |
| .beta.-Narcotine |
| Noscapine (TN) |
| (-)-noscapine |
| NCGC00016388-01 |
| [S-(R*,S*)]-6,7-Dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo[4,5-g]isoquinolin-5-yl)-1(3H)-Isobenzofuranone |
| 08N |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-((5R)-5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo(4,5-g)isoquinolin-5-yl)-, (3S)- |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-[(5R)-5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo[4,5-g]isoquinolin-5-yl]-, (3S)- |
| CAS-128-62-1 |
| Prestwick_959 |
| (-)-a-Narcotine |
| 1-.alpha.-Narcotine |
| Tocris-1697 |
| NOSCAPINE [INN] |
| NOSCAPINE [JAN] |
| NOSCAPINE [MI] |
| NOSCAPINE [HSDB] |
| Prestwick0_000563 |
| Prestwick1_000563 |
| Prestwick2_000563 |
| Prestwick3_000563 |
| NARCOTINUM [HPUS] |
| CBMicro_048259 |
| D0Y8AW |
| NOSCAPINE [MART.] |
| NOSCAPINE [USP-RS] |
| NOSCAPINE [WHO-DD] |
| NOSCAPINE [WHO-IP] |
| SCHEMBL4559 |
| Noscapine [BAN:INN:JAN] |
| (S,R)-Noscapine, 97% |
| Lopac0_000840 |
| BSPBio_000346 |
| MLS000069475 |
| MLS001060855 |
| SPBio_002565 |
| Noscapine (JP15/USP/INN) |
| Noscapine (JP17/USP/INN) |
| BPBio1_000382 |
| CHEMBL364713 |
| NOSCAPINE DL-FORM [MI] |
| NOSCAPINE [EP MONOGRAPH] |
| NOSCAPINE, (+/-)- |
| GTPL10212 |
| NOSCAPINE [USP MONOGRAPH] |
| NOSCAPINUM [WHO-IP LATIN] |
| DTXSID901032089 |
| HMS1569B08 |
| HMS2096B08 |
| HMS2269P05 |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo[4,5-g]isoquinolin-5-yl)-, (R*,S*)-(.+.)- |
| 6,7-Dimethoxy-3-(4-methoxy-6-methyl-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-5-yl)-2-benzofuran-1(3H)-one, (S-(R*,S*))- # |
| Phthalide, 6,7-dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo(4,5-g)isoquinolin-5-yl)- |
| (+/-)-.ALPHA.-NARCOTINE |
| Tox21_110413 |
| BBL012344 |
| BDBM50424716 |
| CB3304 |
| MFCD00069316 |
| NSC121869 |
| STK054401 |
| Noscapine 1.0 mg/ml in Acetonitrile |
| AKOS000278036 |
| Tox21_110413_1 |
| CCG-204096 |
| CS-5115 |
| DB06174 |
| NSC-121869 |
| SDCCGSBI-0048054.P004 |
| NCGC00023230-02 |
| NCGC00023230-04 |
| NCGC00023230-05 |
| NCGC00023230-07 |
| NCGC00023230-08 |
| NCGC00023230-10 |
| NCGC00023230-14 |
| (3S)-6,7-Dimethoxy-3-[(5R)-5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo[4,5-g]isoquinolin-5-yl]-1(3H)-isobenzofuranone |
| AC-20272 |
| AC-33191 |
| HY-13716 |
| NCI60_004322 |
| VS-03291 |
| BIM-0048054.P001 |
| C09592 |
| D01036 |
| EN300-24402818 |
| SR-01000075529-6 |
| W-201012 |
| BRD-K89237706-001-03-8 |
| Q60998699 |
| Noscapine, European Pharmacopoeia (EP) Reference Standard |
| Noscapine, United States Pharmacopeia (USP) Reference Standard |
| WLN: T C566 DO FO KN EH & & TJ HO1 K1 J-DT56 BVO DHJ HO1 IO1 |
| (3S)-3-[(5R)-6-methyl-4-(methyloxy)-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-6,7-bis(methyloxy)-2-benzofuran-1(3H)-one |
| (3S)-6,7-dimethoxy-3-[(5R)-4-methoxy-6-methyl-7,8-dihydro-5H-[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-3H-isobenzofuran-1-one |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-(5,6,7,8- tetrahydro-4-methoxy-6-methyl-1,3-dioxolo[4,5-g]isoquinolin -5-yl)-, [S-(R,S)]- |
| 1(3H)-Isobenzofuranone, 6,7-dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo(4,5-g)isoquinolin-5-yl)-, (R*,S*)-(+-)- |
| 1(3H)-Isobenzofuranone,7-dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo[4,5-g]isoquinolin-5-yl)-, [S-(R*,S*)]- |
| Phthalide,7-dimethoxy-3-(5,6,7,8-tetrahydro-4-methoxy-6-methyl-1,3-dioxolo[4,5-g]isoquinolin-5-yl)- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|