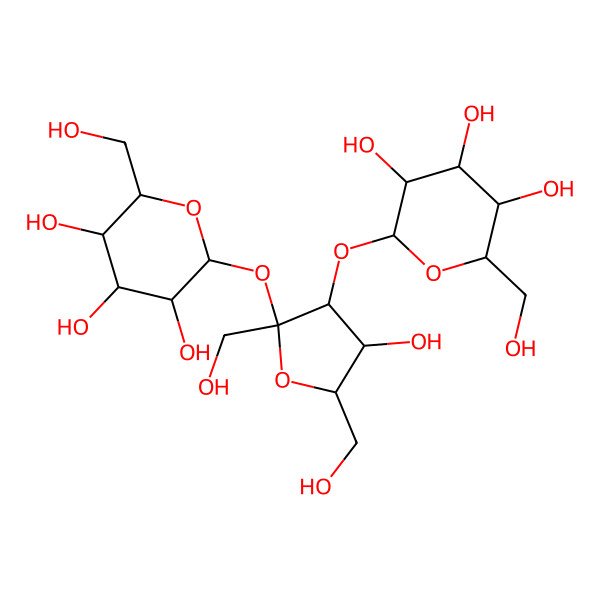
| D-(+)-Melezitose |
| 597-12-6 |
| Melizitose |
| D-Melezitose |
| (+)-Melezitose |
| Melicitose |
| UNII-T4T25QN29L |
| CHEBI:6731 |
| T4T25QN29L |
| NSC 2080 |
| EINECS 209-894-9 |
| AI3-19426 |
| (+)-Melezitose;D-Melezitose |
| (2R,3R,4S,5S,6R)-2-[(2S,3S,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)-2-[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxyoxolan-3-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol |
| D-((addition))-Melezitose |
| NSC-2080 |
| .alpha.-D-Glucopyranoside, O-.alpha.-D-glucopyranosyl-(1->3)-.beta.-D-fructofuranosyl |
| D-(+)Melezitose |
| MELEZITOSE [MI] |
| D(+)-melezitose 1-hydrate |
| MELEZITOSE, (+)- |
| SCHEMBL131977 |
| CHEMBL386007 |
| DTXSID20883458 |
| HY-N2340 |
| s5142 |
| AKOS015955765 |
| CCG-269726 |
| CS-6328 |
| (2R,3R,4S,5S,6R)-2-{[(2S,3S,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)-2-{[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}oxolan-3-yl]oxy}-6-(hydroxymethyl)oxane-3,4,5-triol |
| AS-56726 |
| M0049 |
| C08243 |
| C75789 |
| Q418842 |
| 58079E13-F3CD-4185-B028-4BD36A97D4D3 |
| .alpha.-D-Glucopyranoside, O-.alpha.-D-glucopyranosyl-(1-3)-.beta.-D-fructofuranosyl- |
| alpha-D-Glucopyranoside, O-alpha-D-glucopyranosyl-(1->3)-beta-D-fructofuranosyl |
| alpha-D-Glucopyranoside, O-alpha-D-glucopyranosyl-(1.fwdarw.3)-beta-D-fructofuranosyl |
| alpha-D-glucopyranosyl-(1->3)-beta-D-fructofuranosyl alpha-D-glucopyranoside |
| O-alpha-D-glucopyranosyl-(1,3)-beta-D-fructofuranosyl-alpha-D-glucopyranoside |
| O-alpha-D-glucopyranosyl-(1-3)-beta-D-fructofuranosyl-alpha-D-glucopyranoside |
| O-alpha-D-Glucopyranosyl-(1?3)-beta-D-fructofuranosyl-alpha-D-glucopyranoside |
| WURCS=2.0/2,3,2/[a2122h-1a_1-5][ha122h-2b_2-5]/1-2-1/a1-b2_b3-c1 |
| (2R,2'R,3S,3'S,4S,4'S,5R,5'R,6R,6'R)-6,6'-((2S,3S,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)tetrahydrofuran-2,3-diyl)bis(oxy)bis(2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol) |
| (2R,3R,4S,5S,6R)-2-[(2S,3S,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)-2-[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydropyran-2-yl]oxy-tetrahydrofuran-3-yl]oxy-6-(hydroxymethyl)tetrahydropyran-3,4,5-triol |
| (2R,3R,4S,5S,6R)-2-[(2S,3S,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)-3-[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxyoxolan-2-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol |
| .alpha.-D-Glucopyranoside, O-.alpha.-D-glucopyranosyl-(1.fwdarw.3)-.beta.-D-fructofuranosyl |
| Glucopyranoside, O-.alpha.-D-glucopyranosyl-(1.fwdarw.3)-.beta.-D-fructofuranosyl, .alpha.-D- |
| O-.ALPHA.-D-GLUCOPYRANOSYL-(1->3)-.BETA.-D-FRUCTOFURANOSYL-.ALPHA.-D-GLUCOPYRANOSIDE |
|
There are more than 10 synonyms. If you wish to see them all click here.
|