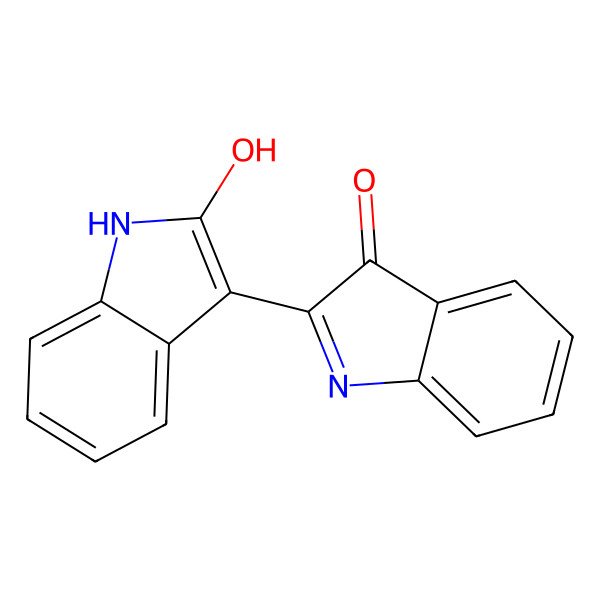
| 479-41-4 |
| Indigo Red |
| Couroupitine B |
| Indigopurpurin |
| 906748-38-7 |
| (Z)-[2,3'-biindolinylidene]-2',3-dione |
| [2,3'-Biindolinylidene]-2',3-dione |
| Isoindirubin |
| Isoindirubine |
| Isoindogotin |
| 397242-72-7 |
| Indirubin [MI] |
| NSC 105327 |
| NSC-105327 |
| (E)-[2,3'-biindolinylidene]-2',3-dione |
| 2H-Indol-2-one, 3-(1,3-dihydro-3-oxo-2H-indol-2-ylidene)-1,3-dihydro- |
| C.I. 73200 |
| Indirubin 3E-form [MI] |
| 3-(3-oxo-1H-indol-2-ylidene)-1H-indol-2-one |
| 1LXW6D3W2Z |
| V86L8P74GI |
| 2-(2-hydroxy-1H-indol-3-yl)indol-3-one |
| C.I. 75790 |
| 3-(1,3-dihydro-3-oxo-2h-indol-2-ylidene)-1,3-dihydro-2h-indol-2-one |
| MFCD00956441 |
| (Z)-[2,3 inverted exclamation mark -Biindolinylidene]-2 inverted exclamation mark ,3-dione |
| (3Z)-3-(1,3-Dihydro-3-oxo-2H-indol-2-ylidene)-1,3-dihydro-2H-indol-2-one |
| (3E)-3-(1,3-Dihydro-3-oxo-2H-indol-2-ylidene)-1,3-dihydro-2H-indol-2-one |
| 2H-Indol-2-one, 3-(1,3-dihydro-3-oxo-2H-indol-2-ylidene)-1,3-dihydro-, (3E)- |
| SMR000466311 |
| BRN 0088279 |
| UNII-V86L8P74GI |
| Indirubin,(S) |
| Indirubin- Bio-X |
| INDARUBICIN |
| (delta2,3'-Biindoline)-2',3-dione |
| CPD000466311 |
| Indirubin derivative, 1 |
| BiomolKI_000069 |
| EM-A05-INDIRUBIN |
| [.DELTA.2,3-dione |
| BiomolKI2_000073 |
| UNII-1LXW6D3W2Z |
| INDIRUBIN [WHO-DD] |
| (3Z)-3-(3-oxoindolin-2-ylidene)indolin-2-one |
| (delta(sup 2,3')-BIINDOLINE)-2',3-DIONE |
| SCHEMBL27678 |
| BMK1-G9 |
| BSPBio_001110 |
| KBioGR_000450 |
| KBioSS_000450 |
| 5-24-08-00507 (Beilstein Handbook Reference) |
| MLS000759416 |
| MLS001424211 |
| MLS002473308 |
| MLS006010732 |
| CHEMBL35349 |
| BDBM7392 |
| CHEMBL259664 |
| SCHEMBL9899338 |
| (3E)-3-(3-oxo-1H-indol-2-ylidene)-1H-indol-2-one |
| CHEMBL1276127 |
| CHEMBL3185783 |
| CHEBI:92322 |
| cid_5318433 |
| KBio2_000450 |
| KBio2_003018 |
| KBio2_005586 |
| KBio3_000839 |
| KBio3_000840 |
| EX-A347 |
| JNLNPCNGMHKCKO-UHFFFAOYSA-N |
| Bio2_000395 |
| Bio2_000875 |
| DTXSID201026053 |
| DTXSID501026052 |
| HMS1362H11 |
| HMS1792H11 |
| HMS1990H11 |
| HMS2051H20 |
| HMS2234G06 |
| HMS3369O15 |
| HMS3393H20 |
| HMS3403H11 |
| HMS3656O13 |
| AMY39890 |
| BCP28869 |
| HY-N0117 |
| AC1180 |
| BDBM50023867 |
| BDBM50349806 |
| FD9058 |
| MFCD00221745 |
| NSC105327 |
| s2386 |
| 2H-Indol-2-one,3-(1,3-dihydro-3-oxo-2H-indol-2-ylidene)-1,3-dihydro- |
| 3-(1,3-dihydro-3-oxo-2h-indol-2-ylidene)-1,3-dihydro-2h-indol-2-on |
| AKOS015895136 |
| AKOS028108775 |
| AKOS032455876 |
| AC-8003 |
| BCP9000788 |
| CCG-100673 |
| CCG-101058 |
| CCG-267073 |
| CS-3682 |
| DB12379 |
| NC00308 |
| SB64698 |
| IDI1_002150 |
| (2'Z)-Indirubin, >=98% (HPLC) |
| NCGC00163356-01 |
| NCGC00163356-02 |
| NCGC00163356-03 |
| NCGC00163356-04 |
| NCGC00179302-02 |
| (Z)-[2,3-Biindolinylidene]-2,3-dione |
| AC-29931 |
| BI164578 |
| CS-12423 |
| LS-14462 |
| PD003207 |
| SMR004701694 |
| SY058396 |
| 3-(3-oxoindolin-2-ylidene)indolin-2-one |
| 3-(3-indolinone-2-ylidene)-indolin-2-one |
| UNM-0000305766 |
| FT-0627199 |
| I0868 |
| SW197688-2 |
| AB00639939-06 |
| 2-(2-oxo-1H-indol-3-ylidene)-1H-indol-3-one |
| A827403 |
| A922429 |
| SR-01000759396 |
| Q-100514 |
| Q1661452 |
| SR-01000759396-5 |
| BRD-K17894950-001-03-6 |
| BRD-K17894950-001-04-4 |
| Q27164070 |
| 3-(3-oxidanylidene-1H-indol-2-ylidene)-1H-indol-2-one |
| 2H-Indol-2-one,3-dihydro-3-oxo-2H-indol-2-ylidene)-1,3-dihydro- |
| (3E)-3-(3-Oxo-1,3-dihydro-2H-indol-2-ylidene)-1,3-dihydro-2H-indo l-2-one |
| (3z)-3-(3-oxo-1,3-dihydro-2h-indol-2-ylidene)-1,3-dihydro-2h-indol-2-one |
| (Z)-[2,3invertedexclamationmark-Biindolinylidene]-2invertedexclamationmark,3-dione |
| 2-[(3Z)-2-oxo-2,3-dihydro-1H-indol-3-ylidene]-2,3-dihydro-1H-indol-3-one |
|
There are more than 10 synonyms. If you wish to see them all click here.
|