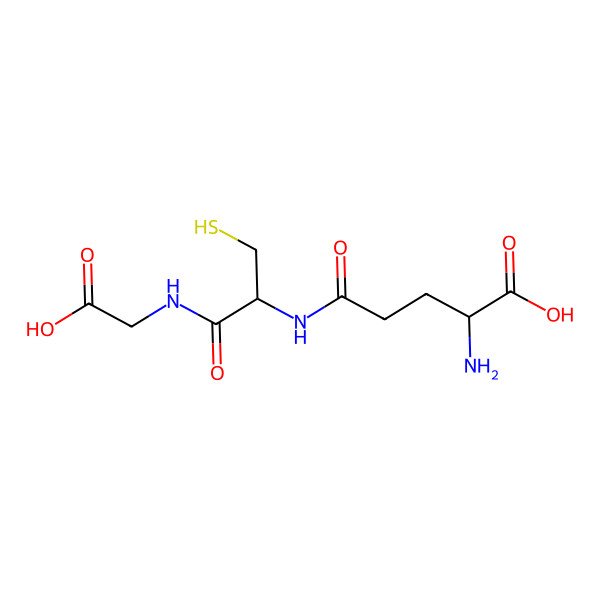
| 70-18-8 |
| L-Glutathione |
| Glutathion |
| Isethion |
| L-Glutathione reduced |
| Tathion |
| Glutathione-SH |
| reduced glutathione |
| Glutinal |
| Tathione |
| Deltathione |
| Neuthion |
| Copren |
| Glutide |
| Triptide |
| Ledac |
| Glutatione |
| GSH |
| Glutatiol |
| Panaron |
| Glutathione SH |
| L-Glutatione |
| Glutathione (reduced) |
| glutathione reduced |
| gamma-L-Glutamyl-L-cysteinylglycine |
| Agifutol S |
| gamma-L-glutamyl-L-cysteinyl-glycine |
| Glutathione [JAN] |
| 5-L-Glutamyl-L-cysteinylglycine |
| Glutham |
| Aec glutathione |
| L-Glutathione, reduced |
| L-gamma-glutamyl-L-cysteinylglycine |
| gamma-L-Glutamylcysteinylglycine |
| Reduced l-glutathione |
| CCRIS 2094 |
| glutathione red |
| red. glutathione |
| Bakezyme RX |
| N-(N-gamma-L-Glutamyl-L-cysteinyl)glycine |
| N-(N-L-gamma-Glutamyl-L-cysteinyl)glycine |
| UNII-GAN16C9B8O |
| EINECS 200-725-4 |
| GAN16C9B8O |
| glycine, N-(N-L-gamma-glutamyl-L-cysteinyl)- |
| NSC 400639 |
| L-Glutathione reduce |
| Glycine, L-gamma-glutamyl-L-cysteinyl- |
| (2S)-2-amino-5-[[(2R)-1-(carboxymethylamino)-1-oxo-3-sulfanylpropan-2-yl]amino]-5-oxopentanoic acid |
| (S)-2-Amino-5-(((R)-1-((carboxymethyl)amino)-3-mercapto-1-oxopropan-2-yl)amino)-5-oxopentanoic acid |
| DTXSID6023101 |
| C10H17N3O6S |
| CHEBI:16856 |
| Glutathione [BAN:JAN] |
| Glutathione (Reduced type) |
| L-glutamyl-L-cysteinylglycine |
| L-Glutathione (reduced form) |
| Glycine, L-.gamma.-glutamyl-L-cysteinyl- |
| DTXCID903101 |
| BenzenaMine, 2-[(4-Methoxyphenyl)Methoxy]- |
| (2S)-2-amino-4-{[(1R)-1-[(carboxymethyl)carbamoyl]-2-sulfanylethyl]carbamoyl}butanoic acid |
| 106272-20-2 |
| 95687-20-0 |
| GLUTATHIONE (II) |
| GLUTATHIONE [II] |
| N-(N-L-.gamma.-Glutamyl-L-cysteinyl)glycine |
| N5-((R)-1-((carboxymethyl)amino)-3-mercapto-1-oxopropan-2-yl)-L-glutamine |
| GLUTATHIONE (MART.) |
| GLUTATHIONE [MART.] |
| GLUTATHIONE (USP-RS) |
| GLUTATHIONE [USP-RS] |
| [Glu(-Cys)]n-Gly |
| GLUTATHIONE (EP MONOGRAPH) |
| GLUTATHIONE [EP MONOGRAPH] |
| MFCD00065939 |
| NSC400639 |
| CAS-70-18-8 |
| (S)-2-Amino-5-((R)-1-(carboxymethylamino)-3-mercapto-1-oxopropan-2-ylamino)-5-oxopentanoic acid |
| Glutathione, Reduced |
| SR-05000002567 |
| gamma L Glu L Cys Gly |
| gamma-L-Glu-L-Cys-Gly |
| L-Glutathione reduced form |
| phytochelatins |
| ReadiSorb |
| Glutathione; |
| 1lbk |
| NCGC00094976-01 |
| gamma L Glutamyl L Cysteinylglycine |
| Tathion (TN) |
| Glutathione (JP17) |
| Spectrum_000419 |
| 1oe7 |
| 1oe8 |
| 1r4w |
| C(N-.gamma.Glu-)G |
| Glycine, N-(N-L-.gamma.-glutamyl-L-cysteinyl)- |
| GLUTATHIONE [MI] |
| Reduced Glutathione,(S) |
| Spectrum2_001500 |
| Spectrum3_000946 |
| Spectrum4_001056 |
| Spectrum5_000940 |
| ?-Glutamylcysteinylglycine |
| Glutathione, Reduced Form |
| GLUTATHIONE [INCI] |
| bmse000185 |
| bmse000952 |
| bmse000956 |
| D02HFD |
| GLUTATHIONE [VANDF] |
| Cys(N-.gamma.Glu-)-Gly |
| SCHEMBL9167 |
| CHEMBL1543 |
| GLUTATHIONE [WHO-DD] |
| KBioGR_001352 |
| KBioSS_000899 |
| MLS001333069 |
| DivK1c_000075 |
| SPECTRUM1502248 |
| SPBio_001519 |
| gamma-glutamyl-cysteinyl-glycine |
| GTPL6737 |
| ?-L-Glutamyl-L-cysteinylglycine |
| L-?-glutamyl-L-cysteinylglycine |
| L-Glutathione reduced, 97.0% |
| CHEBI:60836 |
| HMS500D17 |
| KBio1_000075 |
| KBio2_000899 |
| KBio2_003467 |
| KBio2_006035 |
| KBio3_002012 |
| (gamma-Glutamylcysteine)n-glycine |
| L-g-glutamyl-L-cysteinyl-glycine |
| y-L-Glutamyl-L-cysteinyl-glycine |
| L-?-glutamyl-L-cysteinyl-glycine |
| NINDS_000075 |
| HMS1921N22 |
| Pharmakon1600-01502248 |
| HY-D0187 |
| L-Glutathione reduced, >=98.0% |
| gam.-L-Glutamyl-L-cysteinyl-glycine |
| Tox21_111371 |
| BDBM50422268 |
| C10-H17-N3-O6-S |
| CCG-38876 |
| L-gamma-glutamyl-L-cysteinyl-glycine |
| NSC758199 |
| s4606 |
| AKOS015999135 |
| Tox21_111371_1 |
| CS-7948 |
| DB00143 |
| Glicina, l-gamma-glutamil-l-cisteinil- |
| NSC-758199 |
| SDCCGMLS-0066687.P001 |
| .gamma.-L-Glutamyl-L-cysteinyl-glycine |
| IDI1_000075 |
| N-(N-L-?-Glutamyl-L-cysteinyl)glycine |
| Pharm Biol 11: 539 (1968) |
| SMP1_000247 |
| NCGC00264046-02 |
| DS-14675 |
| GSH;gamma-L-Glutamyl-L-cysteinyl-glycine |
| LS-72665 |
| SMR000857220 |
| SBI-0051743.P002 |
| L-Glutathione reduced, BioXtra, >=98.0% |
| G0074 |
| Glycine, N-(N-L-?-glutamyl-L-cysteinyl)- |
| Glycine, N-(N-L-gamma-glutamyl-L-cysteinyl) |
| C00051 |
| C02471 |
| D00014 |
| EN300-311690 |
| G-3980 |
| P19615 |
| AB00443568_03 |
| Glutathione 100 microg/mL in Acetonitrile:Water |
| A866658 |
| Q116907 |
| SR-05000002567-1 |
| SR-05000002567-2 |
| L-Glutathione reduced, Vetec(TM) reagent grade, >=98% |
| Z2183947556 |
| Glutathione, European Pharmacopoeia (EP) Reference Standard |
| Glutathione, United States Pharmacopeia (USP) Reference Standard |
| Glutathione, Pharmaceutical Secondary Standard; Certified Reference Material |
| L-Glutathione reduced, cell culture tested, BioReagent, >=98.0%, powder |
| (2S)-2-Amino-4-(1-(carboxymethyl)carbamoyl-(2R)-2-sulfanylethylcarbamoyl)butanoic acid |
| (2S)-2-AMINO-4-{[(1R)-1-[(CARBOXYMETHYL)CARBAMOYL]-2-SULFANYLETHYL]CARBAMOYLBUTANOIC ACID |
| (2S)-2-Amino-5-({(2R)-1-[(carboxymethyl)amino]-1-oxo-3-sulfanyl-2-propanyl}amino)-5-oxopentanoic acid |
| glutathione; l-glutathione reduced; 5-l-glutamyl-l-cysteinylglycine; gamma-l-glutamyl-l-cysteinylglycine; gsh |
|
There are more than 10 synonyms. If you wish to see them all click here.
|