| prostaglandin F2alpha |
| 551-11-1 |
| PGF2alpha |
| Prostaglandin F2a |
| amoglandin |
| Enzaprost |
| Protamodin |
| panacelan |
| l-PGF2-alpha |
| Enzaprost F |
| Prostaglandin F2 alpha |
| Prostaglandin F2-alpha |
| Dinoprosta |
| Dinoprostum |
| l-Prostaglandin F2-alpha |
| Prostin F2alpha |
| cyclosin |
| Prostin F2-alpha |
| PGF-2alpha |
| PGF2 alpha |
| PGF2-alpha |
| Prosmon |
| Prostarmon F |
| 9alpha,11alpha-PGF2 |
| PGF2 |
| PGF2a |
| U-14583 |
| CCRIS 4253 |
| Prostaglandin F2.alpha. |
| HSDB 3315 |
| Dinoprostum [INN-Latin] |
| U 14583 |
| Dinoprosta [INN-Spanish] |
| UNII-B7IN85G1HY |
| (+)-Prostaglandin F2alpha |
| B7IN85G1HY |
| Prostin F 2 alpha |
| BRN 2225571 |
| DTXSID9022946 |
| CHEBI:15553 |
| CHEMBL815 |
| U-14,583 |
| 9a,11a-PGF2 |
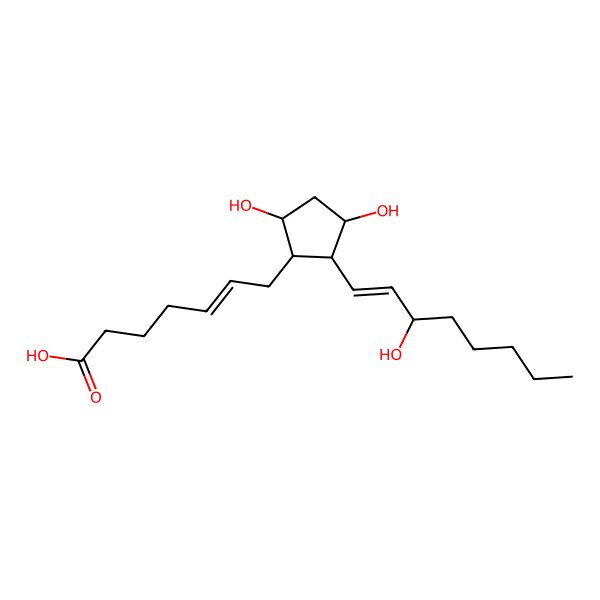
| (Z)-7-[(1R,2R,3R,5S)-3,5-dihydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]cyclopentyl]hept-5-enoic acid |
| DTXCID602946 |
| (+)-Prostaglandin F2a |
| (Z)-7-((1R,2R,3R,5S)-3,5-dihydroxy-2-((S,E)-3-hydroxyoct-1-en-1-yl)cyclopentyl)hept-5-enoic acid |
| 7-(3,5-Dihydroxy-2-(3-hydroxy-1-octenyl)cyclopentyl)-5-heptenoic acid |
| Dinaprost |
| 5-Heptenoic acid, 7-(3,5-dihydroxy-2-(3-hydroxy-1-octenyl)cyclopentyl)-, l- |
| 5-Heptenoic acid, 7-(3,5-dihydroxy-2-(3-hydroxy-1-octenyl)cyclopentyl)-, (1R-(1alpha(Z),2beta(S*,E),3alpha,5alpha))- |
| NCGC00160385-01 |
| Dinoprostum (INN-Latin) |
| Dinoprosta (INN-Spanish) |
| DINOPROST (MART.) |
| DINOPROST [MART.] |
| (5Z,13E,15S)-9alpha,11alpha,15-trihydroxyprosta-5,13-dien-1-oic acid |
| 9S,11R,15S-trihydroxy-5Z,13E-prostadienoic acid |
| (5Z,13E)-(15S)-9alpha,11alpha,15-Trihydroxyprosta-5,13-dienoate |
| PGF2.alpha. |
| Prostaglandin F(sub 2a) |
| 7-[3,5-Dihydroxy-2-(3-hydroxy-1-octenyl)cyclopentyl]-5-heptenoic acid |
| (5Z,9alpha,11alpha,13E,15S)-9,11,15-trihydroxyprosta-5,13-dien-1-oic acid |
| (5Z)-7-[(1R,2R,3R,5S)-3,5-dihydroxy-2-[(1E,3S)-3-hydroxyoct-1-en-1-yl]cyclopentyl]hept-5-enoic acid |
| alpha, PGF2 |
| F2alpha, Prostaglandin |
| 9alpha,11beta PGF2 |
| Dinoprost [USAN:INN:BAN:JAN] |
| (+-)-PGF2alpha |
| SR-01000721847 |
| (E,Z)-(1R,2R,3R,5S)-7-(3,5-Dihydroxy-2-((3S)-(3-hydroxy-1-octenyl))cyclopentyl)-5-heptenoic acid |
| F2 alpha, Prostaglandin |
| (+-)-PGF2-alpha |
| dl-Prostaglandin F2-alpha |
| PGF2-alpha racemic mixt. |
| Prostaglandin F2alpha, dl- |
| (5Z)-7-((1R,2R,3R,5S)-3,5-Dihydroxy-2-((1E,3S)-3-hydroxyoct-1-en-1-yl)cyclopentyl)hept-5-enoic acid |
| 9,11,15-Trihydroxyprosta-5,13-dien-1-oic acid |
| Prosmon (TN) |
| Prosta-5,13-dien-1-oic acid, 9,11,15-trihydroxy-, (5Z,9alpha,11alpha,13E,15S)- |
| Prostaglandin-F2alpha |
| 15-epi PGF2alpha |
| Prostaglandin F2-alpha, racemic mixt. |
| PGF2alpha-[d4] |
| 7-(3, 5-dihydroxy-2-(3-hydroxyl-1-octenyl) cyclopentyl)-5-heptenoic acid |
| BRN 4153898 |
| DINOPROST [INN] |
| DINOPROST [JAN] |
| DINOPROST [HSDB] |
| DINOPROST [USAN] |
| (5Z,9alpha,11alpha,13E,15S)-9,11,15-Trihydroxyprosta-3,13-dien-1-oic acid |
| DINOPROST [WHO-DD] |
| SCHEMBL24292 |
| BSPBio_001494 |
| 23518-25-4 |
| BML1-F08 |
| GTPL1884 |
| Dinoprost (JP17/USAN/INN) |
| 15(R)-Prostaglandin F2 alpha |
| 9,11,15-Trihydroxy-(5Z,9a,11a,13E,15S)-prosta-5,13-dien-1-oic acid |
| EX-A4115A |
| G02AD01 |
| PXGPLTODNUVGFL-YNNPMVKQSA-N |
| HMS1361K16 |
| HMS1791K16 |
| HMS1989K16 |
| HMS2089F11 |
| HMS3402K16 |
| HMS3648N11 |
| (E,Z)-(1R,2R,3R,5S)-7-[3,5-Dihydroxy-2-[(3S)-(3-hydroxy-1-octenyl)]cyclopentyl]-5-heptenoic acid |
| AMY30098 |
| Tox21_111777 |
| BDBM50035622 |
| LMFA03010002 |
| MFCD00135231 |
| PDSP2_000079 |
| AKOS030526160 |
| PROSTAGLANDIN F2.ALPHA. [MI] |
| CCG-208257 |
| CS-4232 |
| DB12789 |
| 5-Heptenoic acid, 7-(3,5-dihydroxy-2-(3-hydroxy-1-octenyl)cyclopentyl)-, (+-)- |
| 5-HEPTENOIC ACID, 7-(3,5-DIHYDROXY-2-(3-HYDROXY-1-OCTENYL)CYCLOPENTYL)-, dl- |
| IDI1_033964 |
| NCGC00160385-02 |
| NCGC00160385-04 |
| NCGC00160385-05 |
| NCGC00160385-06 |
| AS-56892 |
| CAS-551-11-1 |
| HY-12956 |
| P1885 |
| C00639 |
| D00081 |
| A830494 |
| EN300-21689745 |
| Q421375 |
| SR-01000946457 |
| SR-01000721847-2 |
| SR-01000721847-4 |
| SR-01000946457-1 |
| (5Z,9alpha,11alpha,13E,15S)-9,11,15-Trihydroxyprosta-5,13-dienoate |
| (5z,9alpha,11alpha,13e,15s)-9,11,15-trihydroxyprosta-5,13-dienoic acid |
| Prosta-10,13-dien-1-oic acid, 9,11,15-trihydroxy-, (5Z,9.alpha.,11.alpha.,13E,15S) |
| (Z)-7-((1R,2R,3R,5S)-3,5-Dihydroxy-2-((S,E)-3-hydroxyoct-1-enyl)-cyclopentyl)-hept-5-enoic acid |
| (z)-7-[(1r,2r,3r,5s)-3,5-dihydroxy-2-((s)-(e)-3-hydroxyoct-1-enyl)-cyclopentyl]hept-5-enoic acid |
| (Z)-7-[(1R,2R,3R,5S)-3,5-dihydroxy-2-{(E)-(3S)-3-hydroxy-1-octenyl}-cyclopentyl]-5-heptenoic acid |
| 13535-33-6 |
| Prosta-5,13-dien-1-oic acid, 9,11,15-trihydroxy-, (5Z,9-alpha,11-alpha,13E,15S)-(+-)- |
|
There are more than 10 synonyms. If you wish to see them all click here.
|