Crocinervolide
| Internal ID | 8b5864e3-9b5e-4860-a985-ae172bc3da52 |
| Taxonomy | Organoheterocyclic compounds > Lactones > Gamma butyrolactones |
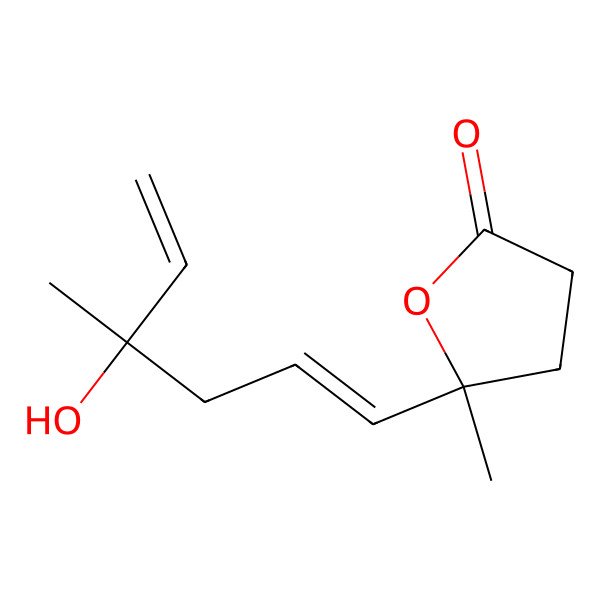
| IUPAC Name | 5-[(1E)-4-hydroxy-4-methylhexa-1,5-dienyl]-5-methyloxolan-2-one |
| SMILES (Canonical) | CC1(CCC(=O)O1)C=CCC(C)(C=C)O |
| SMILES (Isomeric) | CC1(CCC(=O)O1)/C=C/CC(C)(C=C)O |
| InChI | InChI=1S/C12H18O3/c1-4-11(2,14)7-5-8-12(3)9-6-10(13)15-12/h4-5,8,14H,1,6-7,9H2,2-3H3/b8-5+ |
| InChI Key | IQMZMJNVLZXNPZ-VMPITWQZSA-N |
| Popularity | 0 references in papers |
| Molecular Formula | C12H18O3 |
| Molecular Weight | 210.27 g/mol |
| Exact Mass | 210.125594432 g/mol |
| Topological Polar Surface Area (TPSA) | 46.50 Ų |
| XlogP | 1.40 |
| Atomic LogP (AlogP) | 1.97 |
| H-Bond Acceptor | 3 |
| H-Bond Donor | 1 |
| Rotatable Bonds | 4 |
| 125204-36-6 |
| Target | Value | Probability (raw) | Probability (%) |
|---|---|---|---|
| Human Intestinal Absorption | + | 0.9887 | 98.87% |
| Caco-2 | + | 0.8420 | 84.20% |
| Blood Brain Barrier | - | 0.5000 | 50.00% |
| Human oral bioavailability | + | 0.5714 | 57.14% |
| Subcellular localzation | Mitochondria | 0.6038 | 60.38% |
| OATP2B1 inhibitior | - | 0.8582 | 85.82% |
| OATP1B1 inhibitior | + | 0.8754 | 87.54% |
| OATP1B3 inhibitior | + | 0.9119 | 91.19% |
| MATE1 inhibitior | - | 1.0000 | 100.00% |
| OCT2 inhibitior | - | 0.5750 | 57.50% |
| BSEP inhibitior | - | 0.5443 | 54.43% |
| P-glycoprotein inhibitior | - | 0.9737 | 97.37% |
| P-glycoprotein substrate | - | 0.9645 | 96.45% |
| CYP3A4 substrate | + | 0.5988 | 59.88% |
| CYP2C9 substrate | - | 1.0000 | 100.00% |
| CYP2D6 substrate | - | 0.8709 | 87.09% |
| CYP3A4 inhibition | - | 0.7570 | 75.70% |
| CYP2C9 inhibition | - | 0.9235 | 92.35% |
| CYP2C19 inhibition | - | 0.8858 | 88.58% |
| CYP2D6 inhibition | - | 0.9510 | 95.10% |
| CYP1A2 inhibition | - | 0.7941 | 79.41% |
| CYP2C8 inhibition | - | 0.8333 | 83.33% |
| CYP inhibitory promiscuity | - | 0.9734 | 97.34% |
| UGT catelyzed | + | 0.6000 | 60.00% |
| Carcinogenicity (binary) | - | 0.8728 | 87.28% |
| Carcinogenicity (trinary) | Non-required | 0.6717 | 67.17% |
| Eye corrosion | - | 0.9114 | 91.14% |
| Eye irritation | - | 0.5437 | 54.37% |
| Skin irritation | + | 0.6070 | 60.70% |
| Skin corrosion | - | 0.7866 | 78.66% |
| Ames mutagenesis | - | 0.7482 | 74.82% |
| Human Ether-a-go-go-Related Gene inhibition | - | 0.7315 | 73.15% |
| Micronuclear | - | 0.9400 | 94.00% |
| Hepatotoxicity | - | 0.5548 | 55.48% |
| skin sensitisation | + | 0.6248 | 62.48% |
| Respiratory toxicity | + | 0.5111 | 51.11% |
| Reproductive toxicity | - | 0.6544 | 65.44% |
| Mitochondrial toxicity | - | 0.5750 | 57.50% |
| Nephrotoxicity | + | 0.6254 | 62.54% |
| Acute Oral Toxicity (c) | III | 0.7982 | 79.82% |
| Estrogen receptor binding | - | 0.8097 | 80.97% |
| Androgen receptor binding | - | 0.8521 | 85.21% |
| Thyroid receptor binding | - | 0.7524 | 75.24% |
| Glucocorticoid receptor binding | - | 0.5829 | 58.29% |
| Aromatase binding | - | 0.7505 | 75.05% |
| PPAR gamma | - | 0.7084 | 70.84% |
| Honey bee toxicity | - | 0.8768 | 87.68% |
| Biodegradation | - | 0.5250 | 52.50% |
| Crustacea aquatic toxicity | + | 0.5300 | 53.00% |
| Fish aquatic toxicity | + | 0.8434 | 84.34% |
Proven Targets:
| CHEMBL ID | UniProt ID | Name | Min activity | Assay type | Source |
|---|---|---|---|---|---|
| No proven targets yet! | |||||
Predicted Targets (via Super-PRED):
| CHEMBL ID | UniProt ID | Name | Probability | Model accuracy |
|---|---|---|---|---|
| CHEMBL253 | P34972 | Cannabinoid CB2 receptor | 92.82% | 97.25% |
| CHEMBL5619 | P27695 | DNA-(apurinic or apyrimidinic site) lyase | 92.06% | 91.11% |
| CHEMBL5966 | P55899 | IgG receptor FcRn large subunit p51 | 89.93% | 90.93% |
| CHEMBL3108638 | O15164 | Transcription intermediary factor 1-alpha | 88.90% | 95.56% |
| CHEMBL1293249 | Q13887 | Kruppel-like factor 5 | 87.05% | 86.33% |
| CHEMBL3401 | O75469 | Pregnane X receptor | 85.16% | 94.73% |
| CHEMBL2007 | P16234 | Platelet-derived growth factor receptor alpha | 84.15% | 91.07% |
| CHEMBL2581 | P07339 | Cathepsin D | 83.57% | 98.95% |
| CHEMBL3251 | P19838 | Nuclear factor NF-kappa-B p105 subunit | 83.42% | 96.09% |
Below are displayed all the plants proven (via scientific papers) to contain this
compound!
To see more specific details click the taxa you are interested in.
To see more specific details click the taxa you are interested in.
| Conioselinum anthriscoides |