| 57-00-1 |
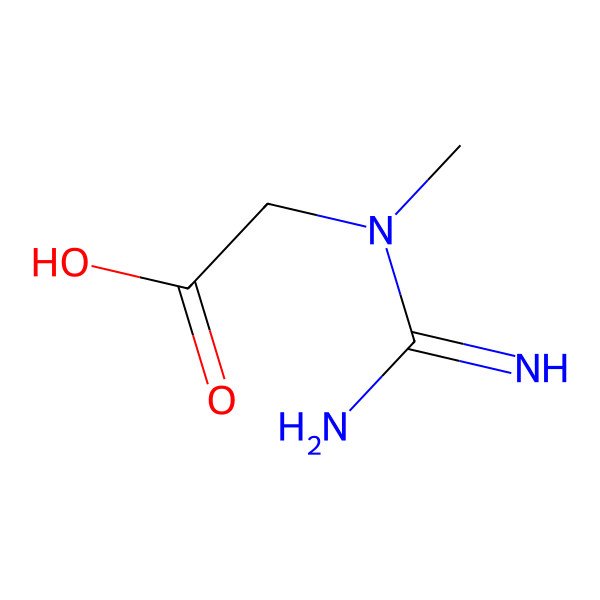
| Creatin |
| Kreatin |
| N-amidinosarcosine |
| Krebiozon |
| N-methyl-N-guanylglycine |
| 2-(1-Methylguanidino)acetic acid |
| methylglycocyamine |
| Creatine, hydrate |
| Glycine, N-(aminoiminomethyl)-N-methyl- |
| Pyrolysate |
| (alpha-Methylguanido)acetic acid |
| Cosmocair C 100 |
| N-carbamimidoyl-N-methylglycine |
| Methylguanidoacetic acid |
| 2-(1-methylcarbamimidamido)acetic acid |
| Kreatinum |
| Neotine |
| Creatine, anhydrous |
| N-(aminoiminomethyl)-N-methylglycine |
| alpha-Methylguanidino acetic acid |
| NSC 8752 |
| NSC-8752 |
| UNII-MU72812GK0 |
| A-MEethyl guanidine acetic acid |
| 2-[carbamimidoyl(methyl)amino]acetic acid |
| CHEBI:16919 |
| AI3-15320 |
| HSDB 7336 |
| MU72812GK0 |
| (.alpha.-Methylguanido)acetic acid |
| (N-methylcarbamimidamido)acetic acid |
| EINECS 200-306-6 |
| ((amino(imino)methyl)(methyl)amino)acetic acid |
| N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE |
| DTXSID1040451 |
| NSC8752 |
| EC 200-306-6 |
| 2-(N-methylcarbamimidamido)acetic acid |
| N-(Aminoiminomethyl)-N-methyl-glycine |
| CREATINE (II) |
| CREATINE [II] |
| CREATINE (USP-RS) |
| CREATINE [USP-RS] |
| (N-(AMIDINO)-N-METHYLAMINO)ACETIC ACID |
| N-(amino(imino)methyl)-N-methylglycine |
| N-[amino(imino)methyl]-N-methylglycine |
| Creatine (8CI) |
| ((amino(imino)methyl)(methyl)amino)acetate |
| [[Amino(imino)methyl](methyl)amino]acetate |
| [[Amino(imino)methyl](methyl)amino]acetic acid |
| N-((E)-AMINO(IMINO)METHYL)-N-METHYLGLYCINE |
| Kre-Alkalyn |
| Methylguanidoacetate |
| IOM |
| MFCD00004282 |
| CREATINE [HSDB] |
| CREATINE [INCI] |
| CREATINE [MI] |
| CREATINE [VANDF] |
| bmse000078 |
| bmse000950 |
| bmse000999 |
| CREATINE [WHO-DD] |
| (alpha-Methylguanido)acetate |
| (a-Methylguanido)acetic acid |
| SCHEMBL21567 |
| Creatine, hydrate (Salt/Mix) |
| CHEMBL283800 |
| GTPL4496 |
| DTXCID9020451 |
| STR05570 |
| BBL012187 |
| BDBM50357229 |
| s5588 |
| STL163516 |
| AKOS005717096 |
| CCG-266112 |
| CS-W011104 |
| DB00148 |
| HY-W010388 |
| Creatine, anhydrous, >=99.0% (NT) |
| 2-(carbamimidoyl-methyl-amino)acetic acid |
| AC-34370 |
| C3610 |
| FT-0624083 |
| C00300 |
| D89574 |
| EN300-1692957 |
| [[Amino(imino)methyl](methyl)amino]acetic acid # |
| Q223600 |
| W-105270 |
| F8880-7830 |
| Z1203577925 |
| 1093737-19-9 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|