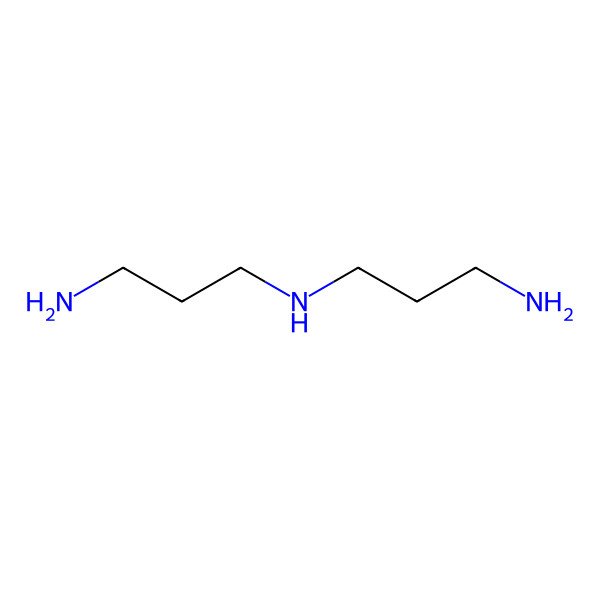
| Bis(3-aminopropyl)amine |
| Dipropylenetriamine |
| 56-18-8 |
| 3,3'-Iminobispropylamine |
| 3,3'-Diaminodipropylamine |
| Caldine |
| 3,3'-Iminobis(propylamine) |
| 1,7-Diamino-4-azaheptane |
| 4-Azaheptane-1,7-diamine |
| N-(3-Aminopropyl)-1,3-propanediamine |
| 3,3-Diaminodipropylamine |
| Aminobis(propylamine) |
| Iminobis(propylamine) |
| 3,3'-Iminodi(propylamine) |
| P 2 (hardener) |
| Imino-bis(3-propylamine) |
| Dipropylentriamin |
| N-3-Aminopropyl-1,3-diaminopropane |
| Dipropylene triamine |
| 1,3-Propanediamine, N-(3-aminopropyl)- |
| 4-Azaheptamethylenediamine |
| Dipropylenetriamine (VAN) |
| N-(3-aminopropyl)propane-1,3-diamine |
| Propylamine, 3,3'-iminobis- |
| Dipropylamine, 3,3'-diamino- |
| N'-(3-aminopropyl)propane-1,3-diamine |
| 3,3'-Iminodipropylamine |
| 1-Propanamine, 3,3'-iminobis- |
| NSC 7773 |
| 1,5,9-triazanonane |
| 3,3'-Iminopropylamine |
| 3,3'-iminobis-Propylamine |
| 3,3'-diamino-Dipropylamine |
| RN1144MGND |
| CHEMBL28743 |
| 1,3-Propanediamine, N1-(3-aminopropyl)- |
| DTXSID0025440 |
| CHEBI:16841 |
| NSC-7773 |
| Initiating explosive iminobispropylamine |
| 3,3'-Iminobis[propylamine] |
| Bis-(3-aminopropyl)amine |
| Dipropylentriamin [German] |
| CCRIS 4826 |
| di(3-aminopropyl)amine |
| EINECS 200-261-2 |
| UN2269 |
| UNII-RN1144MGND |
| BRN 1071254 |
| AI3-25361 |
| Dipropylenetriame |
| N-3-Aminopropyl |
| NSD |
| -1,3-propanediamine |
| 3,3'-Diaminopropylamine |
| Propylamine,3'-iminobis- |
| Dipropylamine,3'-diamino- |
| EC 200-261-2 |
| 1, N-(3-aminopropyl)- |
| 3, 3'-Diaminodipropylamine |
| WLN: Z3M3Z |
| SCHEMBL15387 |
| 1-Propanamine,3'-iminobis- |
| Bis(3,3'-aminopropyl)amine |
| 3,3''-iminodi(propylamine) |
| 4-04-00-01278 (Beilstein Handbook Reference) |
| N,N-bis(3-aminopropyl)amine |
| 3,3''-iminobis(propylamine) |
| 3,3'-iminobis-1-Propanamine |
| 3,3'-azanediyldi(propanamine) |
| DTXCID505440 |
| Bis(3-aminopropyl)amine, 98% |
| 3,3''-azanediyldi(propanamine) |
| DIPROPYLENETRIAMINE [INCI] |
| NSC7773 |
| 3, {3'-Iminobis[propylamine]} |
| N,N-bis-(3-amino-propyl)-amine |
| AMY14287 |
| STR03128 |
| Tox21_202847 |
| BDBM50009367 |
| MFCD00008214 |
| AKOS006223909 |
| UN 2269 |
| CAS-56-18-8 |
| n1-(3-aminopropyl)propane-1,3-diamine |
| N-(3-Aminopropyl)-1, 3-propanediamine |
| NCGC00260393-01 |
| D0090 |
| Initiating explosive iminobispropylamine (dot) |
| 1,3-PROPANEDIAMINE,N-(3-AMINOPROPYL)- |
| C03375 |
| EN300-113765 |
| 3,3'-Iminodipropylamine [UN2269] [Corrosive] |
| Bis(3-aminopropyl)amine, purum, >=97.0% (GC) |
| Q3878510 |
| W-109255 |
| Z1203577926 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|