| 18683-91-5 |
| 107814-37-9 |
| rac-cis-Ambroxol |
| 36557-04-7 |
| Ambroxol Base |
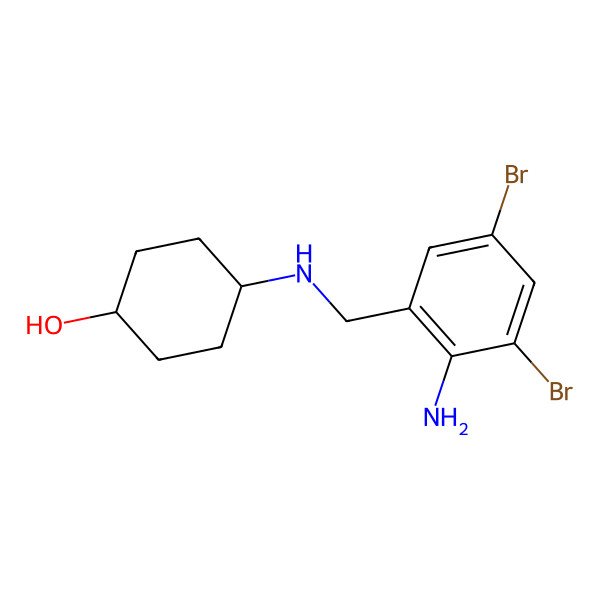
| 4-((2-amino-3,5-dibromobenzyl)amino)cyclohexan-1-ol |
| Ambroxolum |
| Bisolvon metabolite VIII |
| Bromhexine-metabolite VIII |
| NA-872 |
| Tabcin |
| trans-4-((2-Amino-3,5-dibromobenzyl)amino)cyclohexanol |
| Ambroxol, cis- |
| cis-4-((2-Amino-3,5-dibromobenzyl)amino)cyclohexanol |
| Ambroxol [INN] |
| Cyclohexanol, 4-[[(2-amino-3,5-dibromophenyl)methyl]amino]-, trans- |
| 4-[(2-amino-3,5-dibromophenyl)methylamino]cyclohexan-1-ol |
| trans-4-((2-Amino-3,5-dibromobencil)amino)ciclohexanol |
| trans-4-((2-Amino-3,5-dibromobenzyl)amine)cyclohexanol |
| QH6ZT6J071 |
| Cyclohexanol, 4-((2-amino-3,5-dibromobenzyl)amino)- (E)- |
| N-(2-Amino-3,4-dibromocyclohexyl)-trans-4-aminocyclohexanol |
| 4-Hydroxydemethylbromhexine, cis- |
| N-(trans-4-Hydroxycyclohexyl)-(2-amino-3,5-dibromobenzyl)-amine |
| N-(trans-p-Hydroxycyclohexyl)-(2-amino-3,5-dibromobenzyl)amine |
| Ambroxol (INN) |
| Cyclohexanol, 4-(((2-amino-3,5-dibromophenyl)methyl)amino)-, trans- |
| 200168S0CL |
| trans-4-[(2-amino-3,5-dibromobenzyl)amino]cyclohexanol |
| N-(2-Amino-3,4-dibromociclohexil)-trans-4-aminociclohexanol |
| N-(trans-4-Hidroxiciclohexil)-(2-amino-3,5-dibromobencil)amina |
| 1217679-83-8 |
| Ambroxol [INN:BAN] |
| cis-4-(((2-Amino-3,5-dibromophenyl)methyl)amino)cyclohexanol |
| Ambroxolum [INN-Latin] |
| Cyclohexanol, 4-(((2-amino-3,5-dibromophenyl)methyl)amino)-, cis- |
| Cyclohexanol, 4-(((2-amino-3,5-dibromophenyl)methyl)amino)-, trans- (9CI) |
| Bromhexine Metabolite VIII |
| NCGC00016781-04 |
| EINECS 242-500-3 |
| CAS-23828-92-4 |
| Amboxol |
| cis-Ambroxol |
| SR-05000001463 |
| UNII-200168S0CL |
| Tabcin (TN) |
| Spectrum_001346 |
| AMBROXOL [MI] |
| Prestwick0_000366 |
| Prestwick1_000366 |
| Prestwick2_000366 |
| Prestwick3_000366 |
| Spectrum2_001518 |
| Spectrum3_000955 |
| Spectrum4_001068 |
| Spectrum5_001021 |
| trans-4-((2-Amino-3,5-dibromobencil)amino)ciclohexanol [Spanish] |
| N-(2-Amino-3,4-dibromociclohexil)-trans-4-aminociclohexanol [Spanish] |
| AMBROXOL [WHO-DD] |
| EC 242-500-3 |
| N-(trans-4-Hidroxiciclohexil)-(2-amino-3,5-dibromobencil)amina [Spanish] |
| UNII-QH6ZT6J071 |
| Oprea1_766685 |
| SCHEMBL18702 |
| BSPBio_000491 |
| KBioGR_001396 |
| KBioSS_001826 |
| MLS001306470 |
| DivK1c_000027 |
| SCHEMBL423712 |
| SPBio_001595 |
| SPBio_002412 |
| BPBio1_000541 |
| CHEMBL153479 |
| SCHEMBL7855003 |
| 4-[(2-amino-3,5-dibromo-phenyl)methylamino]cyclohexanol |
| CHEMBL1477775 |
| DTXSID8022583 |
| SCHEMBL21765132 |
| BCBcMAP01_000092 |
| CHEBI:92994 |
| GTPL10692 |
| KBio1_000027 |
| KBio2_001826 |
| KBio2_004394 |
| KBio2_006962 |
| KBio3_002050 |
| DTXSID60860228 |
| CHEBI:135590 |
| JBDGDEWWOUBZPM-XYPYZODXSA-N |
| NINDS_000027 |
| 4-{[(2-amino-3,5-dibromophenyl)methyl]amino}cyclohexan-1-ol |
| HMS2089D06 |
| HMS2231B19 |
| HMS3373J22 |
| HMS3886M18 |
| BCP04489 |
| HY-B1039 |
| BDBM50395322 |
| MFCD00242702 |
| MFCD28143339 |
| s5710 |
| AKOS005530704 |
| AKOS015889660 |
| AKOS027338704 |
| AC-8362 |
| Ambroxol hydrochloride impurity D [EP] |
| BCP9000283 |
| CCG-207907 |
| CS-4558 |
| DB06742 |
| SB17463 |
| SB82866 |
| IDI1_000027 |
| SMP1_000014 |
| NCGC00016781-01 |
| NCGC00016781-02 |
| NCGC00016781-08 |
| NCGC00159399-02 |
| NCGC00371077-02 |
| AS-56023 |
| SMR000718792 |
| VS-02240 |
| 4-HYDROXYDEMETHYLBROMHEXINE, TRANS- |
| BCP0726000066 |
| SBI-0051766.P002 |
| AB00514663 |
| CS-0323348 |
| FT-0622261 |
| FT-0630430 |
| FT-0661549 |
| D07442 |
| AB00053639_13 |
| AB01275465-01 |
| A813089 |
| EN300-18524079 |
| Q221637 |
| SR-05000001463-1 |
| AMBROXOL HYDROCHLORIDE IMPURITY D [EP IMPURITY] |
| BRD-K11223672-003-03-7 |
| BRD-K11223672-003-04-5 |
| BRD-K56558538-003-02-8 |
| 4-[(2-amino-3,5-dibromophenyl)methylamino-]cyclohexanol |
| 4-[(2-amino-3,5-dibromophenyl)methylamino]cyclohexanol |
| (1s,4s)-4-((2-amino-3,5-dibromobenzyl)amino)cyclohexanol |
| 4-[(2-Amino-3,5-dibromobenzyl)amino]cyclohexanol, trans- |
| (1r,4r)-4-((2-amino-3,5-dibromobenzyl)amino)cyclohexan-1-ol |
| trans-4-{[(2-amino-3,5-dibromophenyl)methyl]amino}cyclohexanol |
| (1r,4r)-4-{[(2-Amino-3,5-dibromophenyl)methyl]amino}cyclohexan-1-ol |
| Cyclohexanol, 4-[[(2-amino-3,5-dibromo-phenyl)methyl]amino]-, trans |
|
There are more than 10 synonyms. If you wish to see them all click here.
|