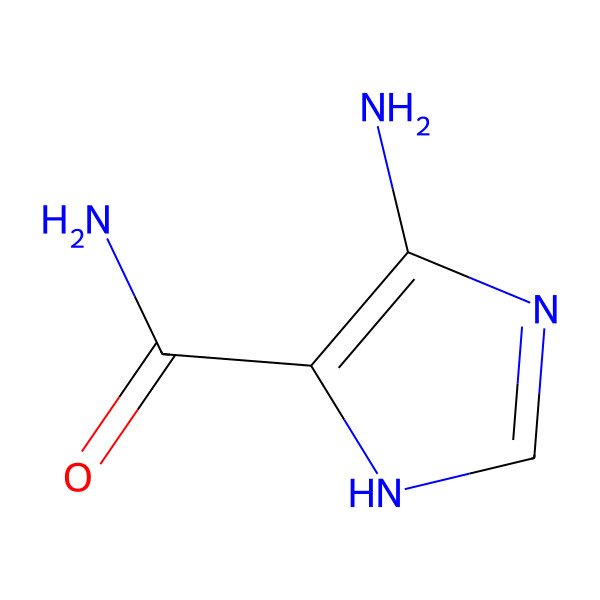
| 5-Aminoimidazole-4-carboxamide |
| 4-Amino-1H-imidazole-5-carboxamide |
| 4-Amino-5-imidazolecarboxamide |
| 5-amino-1H-imidazole-4-carboxamide |
| 5-Amino-4-imidazolecarboxamide |
| AICA |
| 21299-72-9 |
| Colahepat |
| 4-Aminoimidazole-5-carboxamide |
| Aminoimidazole carboxamide |
| 1H-Imidazole-4-carboxamide, 5-amino- |
| 5-amino-3H-imidazole-4-carboxamide |
| Diazol-C |
| 4-Carbamoyl-5-aminoimidazole |
| 5-Aminoimidazol-4-carboxamide |
| 4-Carboxamido-5-aminoimidazole |
| Ba 2756 |
| IMIDAZOLE-4-CARBOXAMIDE, 5-AMINO- |
| 5-amino-imidazole-4-carboxamide |
| 4-Amino-5-imidazole carboxamide |
| 5-Imidazolecarboxamide, 4-amino- |
| 5-Aminoimidazolecarboxamide |
| NSC 7784 |
| 5-Aminoimidazole carboxamide |
| 4-amino-5-carbamoylimidazole |
| AIC |
| NSC-7784 |
| EINECS 206-641-4 |
| UNII-3V0Y2PDE6K |
| 3V0Y2PDE6K |
| CHEBI:2030 |
| IMIDAZOLE-4(OR 5)-CARBOXAMIDE, 5(OR 4)-AMINO- |
| 5-Amino-4-imidazolecarboxyamide |
| 4(5)-Amino-5(4)-Imidazolecarboxamide |
| MFCD02181040 |
| Imidazole-4-carboxamide, 5-amino- (hydrochloride) |
| Dacarbazine impurity B |
| SCHEMBL8024 |
| CHEMBL1610 |
| MLS000701328 |
| Imidazole C-4,5 deriv. 2 |
| WLN: T5M CNJ DVZ EZ |
| BDBM7957 |
| DTXSID8059891 |
| DVNYTAVYBRSTGK-UHFFFAOYSA- |
| NSC7784 |
| Aminoimidazole-4-carboxamide, 5- |
| Imidazole, 5-amino-4-carboxamide |
| HMS2231D11 |
| HMS3369A01 |
| 5-amino-1H-imidazole4-carboxamide |
| BCP22943 |
| 5-amino-1 H-imidazole4-carboxamide |
| 4-azanyl-1H-imidazole-5-carboxamide |
| Imidazole, 4-amino-5-aminocarbonyl- |
| s3651 |
| STL484285 |
| 5-amino-1 H-imidazole-4-carboxamide |
| 4-Amino-5-Imidazolecarboxamide(AICA) |
| 5-Amino-4-imidazolecarboxamide, 95% |
| AKOS009084625 |
| AKOS015855671 |
| 5-Amino-1H-imidazole-4-carboxamide # |
| AC-8311 |
| BA-2756 |
| CCG-266103 |
| CS-W019921 |
| NCGC00245975-01 |
| NCGC00245975-02 |
| AS-12404 |
| HY-41461 |
| LS-78157 |
| SMR000526285 |
| SY009547 |
| AM20090183 |
| AM20090571 |
| DACARBAZINE IMPURITY B [EP IMPURITY] |
| FT-0632037 |
| TEMOZOLOMIDE IMPURITY A [EP IMPURITY] |
| 4-Amino-5-carboxamido imidazole hydrochloride |
| 5-amino-1H-imidazole-4-carboxylic acid amide |
| 5-Amino-3H-imidazole-4-carboxylic acid amide |
| EN300-23736 |
| C04051 |
| O11342 |
| W13853 |
| A815255 |
| A823125 |
| AG-756/20213048 |
| Q-200531 |
| 5-Amino-2-methyl-n-ethyl-n-phenyl-benzenesulfonamide |
| Q27067435 |
| 4-Amino-5-imidazolecarboxamide hydrochloride (Salt/Mix) |
| F0001-1035 |
| 49B332E5-7C8B-4C28-9FDF-05F173231B6A |
| Dacarbazine impurity B, European Pharmacopoeia (EP) Reference Standard |
| TEMOZOLOMIDE IMPURITY, AMINOIMIDAZOLECARBOXAMIDE [USP IMPURITY] |
| InChI=1/C4H6N4O/c5-3-2(4(6)9)7-1-8-3/h1H,5H2,(H2,6,9)(H,7,8) |
| 5AC |
|
There are more than 10 synonyms. If you wish to see them all click here.
|