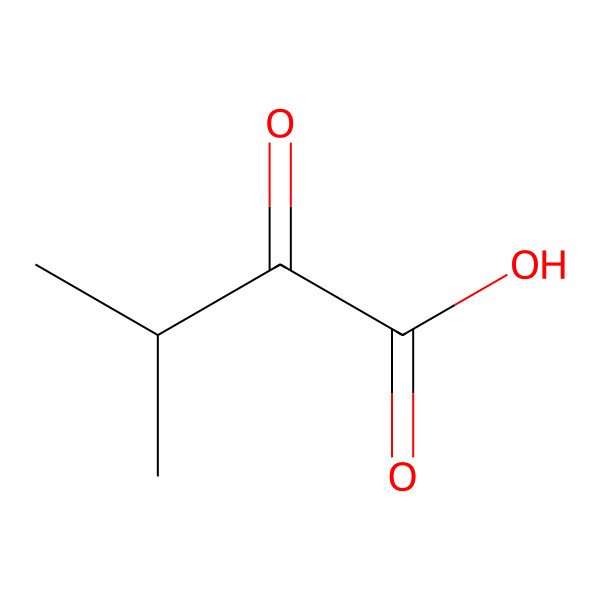
| 759-05-7 |
| 3-methyl-2-oxo-butanoic acid |
| 3-Methyl-2-oxobutyric acid |
| 2-Ketoisovaleric acid |
| alpha-Ketoisovalerate |
| alpha-Ketoisovaleric acid |
| alpha-ketovaline |
| 2-oxoisovalerate |
| 2-Oxoisovaleric acid |
| Butanoic acid, 3-methyl-2-oxo- |
| Dimethylpyruvic acid |
| 2-ketovaline |
| 2-keto-3-methylbutyric acid |
| 3-methyl-2-oxobutanoate |
| Isopropylglyoxylic acid |
| 2-Oxo-3-methylbutanoic acid |
| KETOVALINE |
| 2-oxo-3-methylbutanoate |
| alpha-keto-isovaleric acid |
| alpha-oxoisovalerate |
| alpha-Oxoisovaleric acid |
| 2-oxoisopentanoate |
| 2-Oxo-3-methylbutyric acid |
| alpha-keto-valine |
| 3-Methyl-2-oxobuttersaeure |
| 3-methyl-2-oxo-Butyric acid |
| CHEMBL146554 |
| alpha-oxo-beta-methylbutyricacid |
| UNII-34P71D50E0 |
| CHEBI:16530 |
| (RS)-3-Methyl-2-oxobuttersaeure |
| EINECS 212-065-4 |
| MFCD00040427 |
| 34P71D50E0 |
| a-ketoisovaleric acid |
| 2-Ketoisovalerate |
| KIV |
| Dimethylpyruvate |
| a-Oxoisovalerate |
| a-Ketoisovalerate |
| Isopropylglyoxylate |
| a-keto-Isovalerate |
| a-Oxoisovaleric acid |
| Valine, 2-oxo- |
| dimethyl pyruvic acid |
| 2-Ketoisvaleric acid |
| a-keto-Isovaleric acid |
| a-Oxo-b-methylbutyrate |
| a-keto-b-Methylbutyrate |
| 2-Oxo-3-methylbutyrate |
| 2-keto-3-Methylbutyrate |
| 3-Methyl-2-oxobutinoate |
| 2-Oxo-3-methyl-butyrate |
| 3-methyl-2-oxo-Butyrate |
| 3-methyl-2-oxo-Butanoate |
| a-Oxo-b-methylbutyric acid |
| bmse000101 |
| D03NOU |
| 3-Methyl-2-oxobutanoicacid |
| a-keto-b-Methylbutyric acid |
| SCHEMBL42329 |
| 3-Methyl-2-oxobutinoic acid |
| alpha-Oxo-beta-methylbutyrate |
| 2-OXO-ISOVALERIC ACID |
| alpha-keto-beta-Methylbutyrate |
| 3-methyl-2-oxo butanoic acid |
| DTXSID6061078 |
| alpha-Oxo-beta-methylbutyric acid |
| .ALPHA.-OXOISOVALERIC ACID |
| alpha-keto-beta-Methylbutyric acid |
| .ALPHA.-KETOISOVALERIC ACID |
| FEMA NO. 3869, ACID- |
| .ALPHA.-KETO-ISOVALERIC ACID |
| BDBM50390989 |
| LMFA01020274 |
| s6148 |
| AKOS006272906 |
| CS-W006057 |
| DB04074 |
| DS-0555 |
| HY-W006057 |
| SY005235 |
| 3-METHYL-2-OXOBUTANOIC ACID [FHFI] |
| ALPHA-KETOISOVALERIC ACID; KETOVALINE |
| FT-0616073 |
| .ALPHA.-OXO-.BETA.-METHYLBUTYRIC ACID |
| 3-METHYL-2-OXOBUTYRIC ACIDDISCONTINUED |
| C00141 |
| EN300-142834 |
| Q2823217 |
| W-104383 |
| 9131EA12-7229-4467-B548-60787FD0B557 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|