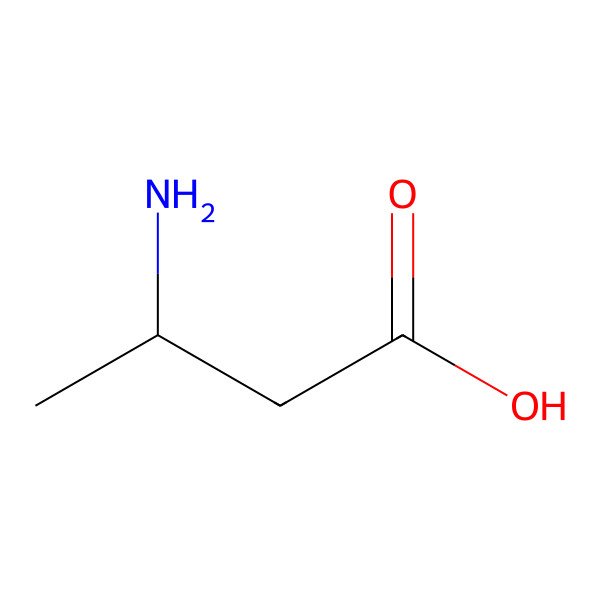
| 541-48-0 |
| DL-3-Aminobutyric acid |
| 3-Aminobutyric acid |
| 2835-82-7 |
| beta-Aminobutyric acid |
| Butanoic acid, 3-amino- |
| DL-3-aminobutanoic acid |
| BABA |
| DL-3-AMINO-N-BUTYRIC ACID |
| DL-beta-homo-alanine |
| Carbocreatine |
| Butyric acid, 3-amino- |
| (+/-)-3-Aminobutyric acid |
| 3-amino-butyric acid |
| 3-methyl-beta-alanine |
| 3-Amino-Butanoic acid |
| .beta.-Aminobutyric acid |
| EINECS 208-783-2 |
| EINECS 220-617-0 |
| 3-aminopent-4-ynoic acid |
| UNII-4282SA5CTS |
| DL-.beta.-Aminobutyric acid |
| (+/-)-3-aminobutanoic acid |
| (3S)-3-Amino-butanoic acid |
| AI3-26821 |
| 4282SA5CTS |
| Butyric acid, 3-amino-, DL- |
| CHEBI:37081 |
| dl-beta-Amino-n-butyric acid |
| MFCD00008087 |
| NSC 77380 |
| NSC-77380 |
| Butanoic acid, 3-amino-, (A+/-)- |
| H-beta-HoAla-OH.HCl |
| DL-beta-Aminobutyric acid |
| R-3-Aminobutyric acid |
| 60625-83-4 |
| b-Homoalanine |
| dl-3-Aminobutyric |
| Tungsten(vi)oxychloride |
| DL-.beta.-Aminobutyrate |
| Dolutegravir Intermediates |
| beta-amino-n-butyric acid |
| .beta.-Aminobutanoic acid |
| 3-Amino-DL-Butyric acid |
| H-DL-Abu(3)-OH |
| DL-Beta-Aminobutyric Acid DL-3-Aminobutyric Acid |
| SCHEMBL44574 |
| .beta.-Amino-n-butyric acid |
| 3-Aminobutanoic acid, 97% |
| Butyric acid, (.+-.)- |
| MLS000069399 |
| (3R)-3-Amino-butanoic acid |
| .beta.-Methyl-.beta.-alanine |
| CHEMBL1995111 |
| dl-.beta.-Amino-n-butyric acid |
| DTXSID4040975 |
| 3-METHYL-.BETA.-ALANINE |
| 3-Amino-(.+-.)-Butyric acid |
| 3-Amino-(.+-.)-Butanoic acid |
| (RS)-3-AMINOBUTANOIC ACID |
| (.+-.)-.beta.-Aminobutyric acid |
| NSC77380 |
| BUTYRIC ACID, .BETA.-AMINO- |
| AKOS005174346 |
| Butanoic acid, 3-amino-, (.+.)- |
| .BETA.-AMINOBUTYRIC ACID [MI] |
| AB87426 |
| AB87563 |
| Butyric acid, 3-amino-, (.+-.)- |
| CS-W009549 |
| HY-W008833 |
| (+/-)-3-AMINO-N-BUTYRIC ACID |
| (+/-)-.BETA.-AMINOBUTYRIC ACID |
| 167222-96-0 |
| AS-11703 |
| NCI60_041705 |
| SMR000058404 |
| SY019803 |
| SY022577 |
| SY022578 |
| (+)-DimEt 2,3-O-isopropylideneD-tartrate |
| LS-182543 |
| .BETA.-AMINOBUTYRIC ACID, (+/-)- |
| A0281 |
| A5389 |
| AM20100428 |
| FT-0625401 |
| FT-0650753 |
| FT-0650754 |
| FT-0689095 |
| EN300-23041 |
| A-5230 |
| A829994 |
| J-502211 |
| J-640366 |
| J-800369 |
| Q3604567 |
| 3-Aminobutanoic acid, Vetec(TM) reagent grade, 97% |
| Z147647144 |
| 4-amino-5-(2-methoxyphenyl)-2H-1,2,4-triazole-3-thione |
| 0B9FE31A-1286-4B2B-B64E-4A3871898B12 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|