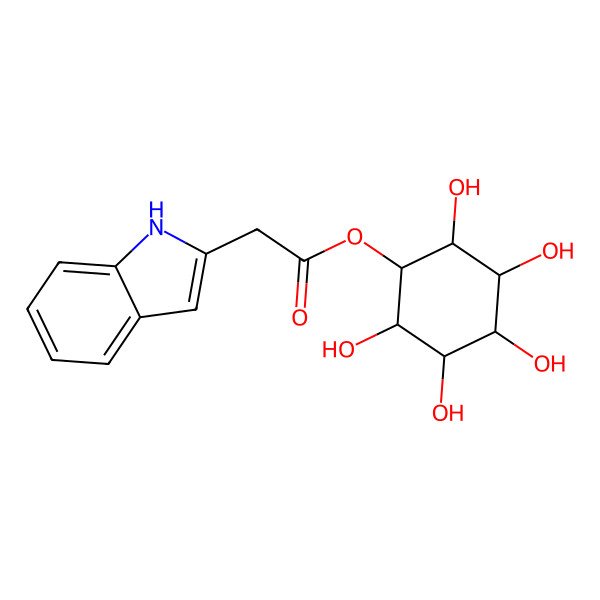
(2,3,4,5,6-pentahydroxycyclohexyl) 2-(1H-indol-2-yl)acetate
| Internal ID | db3b7afa-ea7b-4c13-b0ac-f69fb449b9d8 |
| Taxonomy | Organoheterocyclic compounds > Indoles and derivatives > Indolyl carboxylic acids and derivatives |
| IUPAC Name | (2,3,4,5,6-pentahydroxycyclohexyl) 2-(1H-indol-2-yl)acetate |
| SMILES (Canonical) | C1=CC=C2C(=C1)C=C(N2)CC(=O)OC3C(C(C(C(C3O)O)O)O)O |
| SMILES (Isomeric) | C1=CC=C2C(=C1)C=C(N2)CC(=O)OC3C(C(C(C(C3O)O)O)O)O |
| InChI | InChI=1S/C16H19NO7/c18-10(6-8-5-7-3-1-2-4-9(7)17-8)24-16-14(22)12(20)11(19)13(21)15(16)23/h1-5,11-17,19-23H,6H2 |
| InChI Key | XFLNBCHAOIUSBU-UHFFFAOYSA-N |
| Popularity | 1 reference in papers |
| Molecular Formula | C16H19NO7 |
| Molecular Weight | 337.32 g/mol |
| Exact Mass | 337.11615195 g/mol |
| Topological Polar Surface Area (TPSA) | 143.00 Ų |
| XlogP | -1.40 |
| Atomic LogP (AlogP) | -1.56 |
| H-Bond Acceptor | 7 |
| H-Bond Donor | 6 |
| Rotatable Bonds | 3 |
| There are no found synonyms. |
| Target | Value | Probability (raw) | Probability (%) |
|---|---|---|---|
| Human Intestinal Absorption | + | 0.9681 | 96.81% |
| Caco-2 | - | 0.7922 | 79.22% |
| Blood Brain Barrier | + | 0.6250 | 62.50% |
| Human oral bioavailability | + | 0.5714 | 57.14% |
| Subcellular localzation | Mitochondria | 0.4090 | 40.90% |
| OATP2B1 inhibitior | - | 1.0000 | 100.00% |
| OATP1B1 inhibitior | + | 0.9156 | 91.56% |
| OATP1B3 inhibitior | + | 0.9178 | 91.78% |
| MATE1 inhibitior | - | 0.9400 | 94.00% |
| OCT2 inhibitior | - | 0.9250 | 92.50% |
| BSEP inhibitior | - | 0.8633 | 86.33% |
| P-glycoprotein inhibitior | - | 0.8860 | 88.60% |
| P-glycoprotein substrate | - | 0.9511 | 95.11% |
| CYP3A4 substrate | - | 0.5202 | 52.02% |
| CYP2C9 substrate | - | 0.8202 | 82.02% |
| CYP2D6 substrate | - | 0.7505 | 75.05% |
| CYP3A4 inhibition | - | 0.8538 | 85.38% |
| CYP2C9 inhibition | - | 0.8982 | 89.82% |
| CYP2C19 inhibition | - | 0.8758 | 87.58% |
| CYP2D6 inhibition | - | 0.7872 | 78.72% |
| CYP1A2 inhibition | - | 0.6665 | 66.65% |
| CYP2C8 inhibition | - | 0.5764 | 57.64% |
| CYP inhibitory promiscuity | - | 0.6791 | 67.91% |
| UGT catelyzed | + | 0.8000 | 80.00% |
| Carcinogenicity (binary) | - | 0.9000 | 90.00% |
| Carcinogenicity (trinary) | Non-required | 0.5605 | 56.05% |
| Eye corrosion | - | 0.9933 | 99.33% |
| Eye irritation | - | 0.9121 | 91.21% |
| Skin irritation | - | 0.8189 | 81.89% |
| Skin corrosion | - | 0.9629 | 96.29% |
| Ames mutagenesis | - | 0.5800 | 58.00% |
| Human Ether-a-go-go-Related Gene inhibition | - | 0.7513 | 75.13% |
| Micronuclear | + | 0.7759 | 77.59% |
| Hepatotoxicity | - | 0.6476 | 64.76% |
| skin sensitisation | - | 0.8725 | 87.25% |
| Respiratory toxicity | + | 0.5556 | 55.56% |
| Reproductive toxicity | + | 0.8222 | 82.22% |
| Mitochondrial toxicity | + | 0.7500 | 75.00% |
| Nephrotoxicity | - | 0.7367 | 73.67% |
| Acute Oral Toxicity (c) | III | 0.5284 | 52.84% |
| Estrogen receptor binding | - | 0.5262 | 52.62% |
| Androgen receptor binding | - | 0.6874 | 68.74% |
| Thyroid receptor binding | - | 0.5000 | 50.00% |
| Glucocorticoid receptor binding | + | 0.7246 | 72.46% |
| Aromatase binding | + | 0.6611 | 66.11% |
| PPAR gamma | - | 0.4925 | 49.25% |
| Honey bee toxicity | - | 0.9274 | 92.74% |
| Biodegradation | - | 0.7500 | 75.00% |
| Crustacea aquatic toxicity | - | 0.6600 | 66.00% |
| Fish aquatic toxicity | - | 0.5123 | 51.23% |
Proven Targets:
| CHEMBL ID | UniProt ID | Name | Min activity | Assay type | Source |
|---|---|---|---|---|---|
| No proven targets yet! | |||||
Predicted Targets (via Super-PRED):
| CHEMBL ID | UniProt ID | Name | Probability | Model accuracy |
|---|---|---|---|---|
| CHEMBL5619 | P27695 | DNA-(apurinic or apyrimidinic site) lyase | 95.88% | 91.11% |
| CHEMBL3251 | P19838 | Nuclear factor NF-kappa-B p105 subunit | 93.97% | 96.09% |
| CHEMBL2288 | Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | 90.41% | 91.71% |
| CHEMBL2581 | P07339 | Cathepsin D | 89.94% | 98.95% |
| CHEMBL2035 | P08912 | Muscarinic acetylcholine receptor M5 | 89.44% | 94.62% |
| CHEMBL2563 | Q9UQL6 | Histone deacetylase 5 | 87.76% | 89.67% |
| CHEMBL3108638 | O15164 | Transcription intermediary factor 1-alpha | 86.94% | 95.56% |
| CHEMBL216 | P11229 | Muscarinic acetylcholine receptor M1 | 86.71% | 94.23% |
| CHEMBL3060 | Q9Y345 | Glycine transporter 2 | 84.25% | 99.17% |
| CHEMBL4026 | P40763 | Signal transducer and activator of transcription 3 | 83.03% | 82.69% |
| CHEMBL245 | P20309 | Muscarinic acetylcholine receptor M3 | 82.79% | 97.53% |
| CHEMBL4793 | Q86TI2 | Dipeptidyl peptidase IX | 80.09% | 96.95% |
| PubChem | 77226816 |
| LOTUS | LTS0109219 |
| wikiData | Q105327088 |