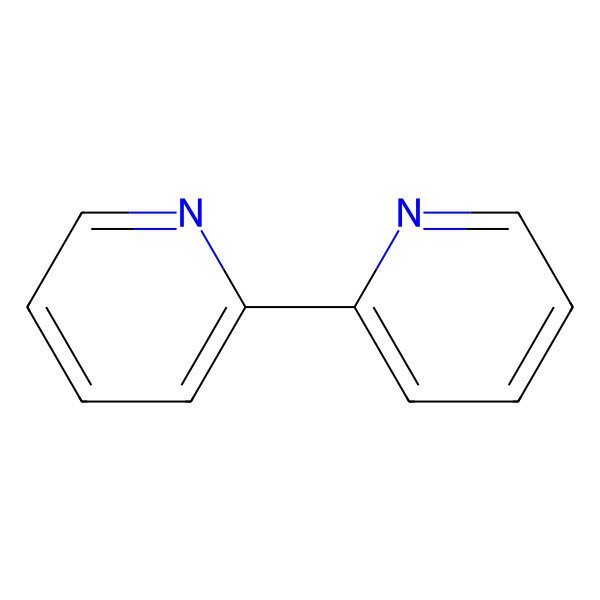
| 366-18-7 |
| 2,2'-Dipyridyl |
| 2,2'-Bipyridyl |
| Bipyridine |
| 2,2'-Dipyridine |
| 2-(2-Pyridyl)pyridine |
| 2,2'-Bipyridin |
| alpha,alpha'-Dipyridyl |
| alpha,alpha'-Bipyridyl |
| dipyridyl |
| Bipyridyl |
| alpha,alpha'-Dipyridine |
| AA-DP |
| 2,2-bipyridyl |
| [2,2]bipyridinyl |
| 2-(pyridin-2-yl)pyridine |
| NSC 1550 |
| .alpha.,.alpha.'-Bipyridine |
| NSC 615009 |
| alpha,alpha'-Bipyridine |
| MFCD00006212 |
| Umdipyridyl |
| .alpha.,.alpha.'-Bipyridyl |
| .alpha.,.alpha.'-Dipyridyl |
| [2,2']BIPYRIDINYL |
| .alpha.,.alpha.'-Dipyridine |
| CHEMBL39879 |
| MLS000069417 |
| DTXSID9040635 |
| CHEBI:30351 |
| 551W113ZEP |
| NSC-1550 |
| CI 588 |
| NSC615009 |
| NSC-615009 |
| BPY |
| SMR000059069 |
| 2,2'-Bipyridine, 99% |
| 2,2-Bipyridine |
| Bipy |
| 2,2 Bipyridyl |
| 2,2-Dipyridyl |
| 2,2' Bipyridine |
| CCRIS 3426 |
| HSDB 5423 |
| alpha,alpha'-Dwupirydylu [Polish] |
| alpha,alpha'-Dwupirydylu |
| EINECS 206-674-4 |
| CI-588 |
| BRN 0113089 |
| bi-pyridine |
| UNII-551W113ZEP |
| AI3-00491 |
| 2-pyridylpyridine |
| 2,2bipyridyl |
| 2'2-bipyridine |
| 2,2'Bipyridine |
| 0BP |
| 2,2' bipyridyl |
| [2,2']bipyridyl |
| 2,2''-bipyridyl |
| 2,2''-dipyridyl |
| 2,2''-bipyridine |
| 2,2''-dipyridine |
| 2,2'- bipyridine |
| 2,2''-Bipyridin |
| 2-pyridin-2-ylpyridin |
| alpha,alpha''-bipyridyl |
| alpha,alpha''-dipyridyl |
| alpha,alpha''-bipyridine |
| alpha,alpha''-dipyridine |
| Maybridge3_006205 |
| Opera_ID_1615 |
| Lopac-D-7505 |
| 2,2'-Bipyridine, ACS |
| EC 206-674-4 |
| SCHEMBL5922 |
| UPCMLD00WV-71 |
| Lopac0_000471 |
| 5-23-08-00016 (Beilstein Handbook Reference) |
| 2,2'-Dipyridyl, ACS grade |
| WLN: T6NJ B- BT6NJ |
| DTXCID7020635 |
| YSSJ00536 |
| 2,2'-BIPYRIDINE [MI] |
| NSC1550 |
| 2,2'-BIPYRIDINE [HSDB] |
| HMS1448K01 |
| HMS2234F20 |
| HMS3261O04 |
| HMS3371D05 |
| BCP27263 |
| HY-D0020 |
| STR02551 |
| Tox21_301430 |
| Tox21_500471 |
| BDBM50042874 |
| CCG-54708 |
| AKOS004901459 |
| 2,2'-Bipyridyl, p.a., 99.5% |
| AC-7556 |
| AM81312 |
| CS-W009134 |
| FS-1056 |
| LP00471 |
| SC11754 |
| SDCCGSBI-0050456.P002 |
| IDI1_017592 |
| NCGC00015364-01 |
| NCGC00015364-02 |
| NCGC00015364-03 |
| NCGC00015364-04 |
| NCGC00015364-05 |
| NCGC00015364-07 |
| NCGC00093368-02 |
| NCGC00093368-03 |
| NCGC00255575-01 |
| NCGC00261156-01 |
| 2,2'-Bipyridyl, >=98.0% (NT) |
| BP-10293 |
| CAS-366-18-7 |
| 2,2'-Bipyridyl, ReagentPlus(R), >=99% |
| B0468 |
| EU-0100471 |
| FT-0632048 |
| FT-0636412 |
| FT-0637152 |
| EN300-52307 |
| A15530 |
| D 7505 |
| D-7200 |
| D-7250 |
| D-7255 |
| 2,2'-Dipyridyl, JIS special grade, >=99.0% |
| 2,2'-Bipyridyl, Vetec(TM) reagent grade, 98% |
| Q209143 |
| SR-01000075829 |
| 2,2'-Bipyridyl, PESTANAL(R), analytical standard |
| SR-01000075829-1 |
| SR-01000075829-3 |
| Z57160161 |
| F0001-1045 |
| 2,2'-Bipyridine;2-(pyridin-2-yl)pyridine;2,2'-Bipyridine |
| 2,2'-Bipyridyl, anhydrous, free-flowing, Redi-Dri(TM), ReagentPlus(R), 99% |
| InChI=1/C10H8N2/c1-3-7-11-9(5-1)10-6-2-4-8-12-10/h1-8 |
|
There are more than 10 synonyms. If you wish to see them all click here.
|