| DPNH |
| 58-68-4 |
| beta-DPNH |
| beta-NADH |
| 1,4-Dihydronicotinamide adenine dinucleotide |
| Dihydrocozymase |
| Coenzyme I, reduced |
| Cozymase I, reduced |
| NADH2 |
| dihydronicotinamide adenine dinucleotide |
| Dihydrocodehydrogenase I |
| Reduced codehydrogenase I |
| Reduced nicotinamide adenine diphosphate |
| Codehydrase I, reduced |
| Dihydronicotinamide mononucleotide |
| Reduced nicotinamide-adenine dinucleotide |
| Codehydrogenase I, reduced |
| Reduced diphosphopyridine nucleotide |
| NAD reduced form |
| Reduced Nicotinamide Adenine Dinucleotide |
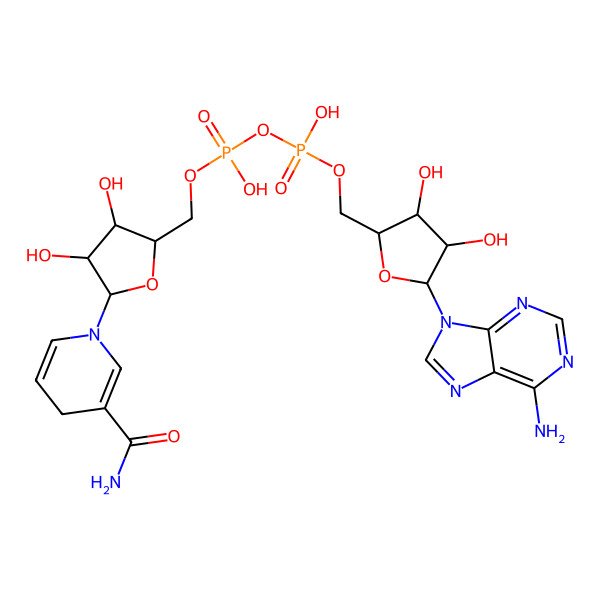
| [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl] [(2R,3S,4R,5R)-5-(3-carbamoyl-4H-pyridin-1-yl)-3,4-dihydroxyoxolan-2-yl]methyl hydrogen phosphate |
| CHEBI:16908 |
| Dihydronicotinamide-adenine dinucleotide |
| 4J24DQ0916 |
| Nicotinamide-adenine dinucleotide, reduced |
| nicotinamide adenine dinucleotide (reduced) |
| Adenosine 5'-(trihydrogen diphosphate), P'.fwdarw.5'-ester with 1,4-dihydro-1-.beta.-D-ribofuranosyl-3-pyridinecarboxamide |
| Adenosine 5'-(trihydrogen pyrophosphate), 5'-5'-ester with 1,4-dihydro-1beta-D-ribofuranosylnicotinamide |
| nadh hydride |
| coenzyme-I |
| b-dpnh |
| UNII-4J24DQ0916 |
| EINECS 200-393-0 |
| ENADA |
| b-NADH |
| Nicotinamide - adenine dinucleotide, reduced |
| NADH [WHO-DD] |
| bmse000054 |
| SCHEMBL8187 |
| NAD REDUCED FORM [MI] |
| GTPL4487 |
| CHEMBL1234616 |
| DTXSID30889320 |
| BOPGDPNILDQYTO-NNYOXOHSSA-N |
| DB00157 |
| [(2R,3S,4R,5R)-5-(6-Aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-[[(2R,3S,4R,5R)-5-(3-carbamoyl-4H-pyridin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-phosphinic acid |
| Adenosine 5'-(trihydrogen diphosphate), P'->5'-ester with 1,4-dihydro-1-beta-D-ribofuranosyl-3-pyridinecarboxamide (9CI) |
| Adenosine 5'-(trihydrogen pyrophosphate), 5'->5'-ester with 1,4-dihydro-1-beta-D-ribofuranosylnicotinamide (8CI) |
| adenosine 5'-{3-[1-(3-carbamoyl-1,4-dihydropyridin-1-yl)-1,4-anhydro-D-ribitol-5-yl] dihydrogen diphosphate} |
| Adenosine pyrophosphate, 5'->5'-ester with 1,4-dihydro-1-beta-D-ribofuranosylnicotinamide (7CI) |
| C00004 |
| EN300-19742425 |
| Q26987453 |
| ((2R,3S,4R,5R)-5-(6-Aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl)methoxy-((((2R,3S,4R,5R)-5-(3-carbamoyl-4H-pyridin-1-yl)-3,4-dihydroxy-oxolan-2-yl)methoxy)hydroxyphosphoryl)oxyphosphinic acid |
| [({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4R,5R)-5-(3-carbamoyl-1,4-dihydropyridin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy})phosphinic acid |
| [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [[(2R,3S,4R,5R)-5-(3-carbamoyl-4H-pyridin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl] hydrogen phosphate |
| Adenosine 5'-(trihydrogen diphosphate), 5'->'-ester with 1,4-dihydro-1-beta-D-ribofuranosyl-3-pyridinecarboxamide |
| Adenosine 5'-(trihydrogen diphosphate), P'-->5'-ester with 1,4-dihydro-1-.beta.-D-ribofuranosyl-3-pyridinecarboxamide |
| ADENOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), P'->5'-ESTER WITH 1,4-DIHYDRO-1-.BETA.-D-RIBOFURANOSYL-3-PYRIDINECARBOXAMIDE |
| Adenosine 5'-(trihydrogen diphosphate), P'->5'-ester with 1,4-dihydro-1-beta-D-ribofuranosyl-3-pyridinecarboxamide |
| Adenosine 5'-(trihydrogen diphosphate), P'.fwdarw.5'-ester with 1,4-dihydro-1-beta-D-ribofuranosyl-3-pyridinecarboxamide |
|
There are more than 10 synonyms. If you wish to see them all click here.
|